Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Modelling ligand selectivity of serine proteases using integrative proteochemometric approaches improves model performance and allows the multi-target dependent interpretation of features†
Qurrat U.
Ain
a,
Oscar
Méndez-Lucio
a,
Isidro Cortés
Ciriano
b,
Thérèse
Malliavin
b,
Gerard J. P.
van Westen
 c and
Andreas
Bender
*a
c and
Andreas
Bender
*a
aCentre for Molecular Informatics, Department of Chemistry, Lensfield Road, CB2 1EW, University of Cambridge, UK. E-mail: ab454@cam.ac.uk
bUnité de Bioinformatique Structurale, Institut Pasteur and CNRS UMR 3825, Structural Biology and Chemistry Department, 25-28, rue du Dr. Roux, 75 724 Paris, France. E-mail: therese.malliavin@pasteur.fr
cEuropean Molecular Biology Laboratory European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK. E-mail: gerardvw@ebi.ac.uk
First published on 16th September 2014
Abstract
Serine proteases, implicated in important physiological functions, have a high intra-family similarity, which leads to unwanted off-target effects of inhibitors with insufficient selectivity. However, the availability of sequence and structure data has now made it possible to develop approaches to design pharmacological agents that can discriminate successfully between their related binding sites. In this study, we have quantified the relationship between 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 625 distinct protease inhibitors and their bioactivity against 67 targets of the serine protease family (20
625 distinct protease inhibitors and their bioactivity against 67 targets of the serine protease family (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 213 data points) in an integrative manner, using proteochemometric modelling (PCM). The benchmarking of 21 different target descriptors motivated the usage of specific binding pocket amino acid descriptors, which helped in the identification of active site residues and selective compound chemotypes affecting compound affinity and selectivity. PCM models performed better than alternative approaches (models trained using exclusively compound descriptors on all available data, QSAR) employed for comparison with R2/RMSE values of 0.64 ± 0.23/0.66 ± 0.20 vs. 0.35 ± 0.27/1.05 ± 0.27 log units, respectively. Moreover, the interpretation of the PCM model singled out various chemical substructures responsible for bioactivity and selectivity towards particular proteases (thrombin, trypsin and coagulation factor 10) in agreement with the literature. For instance, absence of a tertiary sulphonamide was identified to be responsible for decreased selective activity (by on average 0.27 ± 0.65 pChEMBL units) on FA10. Among the binding pocket residues, the amino acids (arginine, leucine and tyrosine) at positions 35, 39, 60, 93, 140 and 207 were observed as key contributing residues for selective affinity on these three targets.
213 data points) in an integrative manner, using proteochemometric modelling (PCM). The benchmarking of 21 different target descriptors motivated the usage of specific binding pocket amino acid descriptors, which helped in the identification of active site residues and selective compound chemotypes affecting compound affinity and selectivity. PCM models performed better than alternative approaches (models trained using exclusively compound descriptors on all available data, QSAR) employed for comparison with R2/RMSE values of 0.64 ± 0.23/0.66 ± 0.20 vs. 0.35 ± 0.27/1.05 ± 0.27 log units, respectively. Moreover, the interpretation of the PCM model singled out various chemical substructures responsible for bioactivity and selectivity towards particular proteases (thrombin, trypsin and coagulation factor 10) in agreement with the literature. For instance, absence of a tertiary sulphonamide was identified to be responsible for decreased selective activity (by on average 0.27 ± 0.65 pChEMBL units) on FA10. Among the binding pocket residues, the amino acids (arginine, leucine and tyrosine) at positions 35, 39, 60, 93, 140 and 207 were observed as key contributing residues for selective affinity on these three targets.
Background
While the human genome encodes more than 3000 potential drug targets,1 only ∼800 of them have been successfully exploited pharmacologically due to a number of limitations (for instance, less compounds satisfying the Lipinski's rule-of-five or the redundancy of targets due to orthologs).2,3 The traditional drug discovery process includes target identification and validation. Subsequent screening campaigns identify hit compounds, which can be optimized to leads and progress into clinical trials.4In silico approaches have been proven to be successful in many phases of this process.5,6It has been recently demonstrated that drugs exert their therapeutic effect by modulating more than one target,7 extending the notion of the one drug one target premise.8 While drug discovery efforts have been long focussed on single target potency optimization,9 the rapid growth of bioactivity databases sparks novel methods to use the data for addressing ligand selectivity and polypharmacology.10–12
Quantitative Structure Activity Relationship (QSAR) modelling relates compound activity on a target to compound properties through machine learning models. However, a single QSAR cannot predict drug potency and selectivity against a panel of targets.13 Current chemogenomic approaches relate biomolecular targets and their ligands on the basis of molecular similarity, where ‘similar ligands show similar activity’ and ‘similar proteins bind similar ligands’.14 Although these techniques enable the extrapolation on either the biological or the chemical side, it is not possible to fully extrapolate the bioactivity of novel compounds on novel targets. Proteochemometric modelling (PCM) integrates compound and target information simultaneously in a single machine learning models. This combination of different, yet complementary, sources of information sets PCM apart from QSAR and chemogenomics, and permits to both inter- and extrapolates the bioactivity of (novel) compounds to (novel) targets.
Technically, the difference between PCM and QSAR is the addition of explicit target descriptors. Thus, each ligand–target interaction is numerically encoded by the concatenation of ligand and target descriptors. Encoding protein information into numerical descriptors is an intricate task.15–17 The most common approaches consist of concatenating amino acid descriptors,15–19 which can correspond to the residues in a binding site or describing the full protein sequence with e.g. the frequency of amino acid pairs, or of exploiting 3-dimensional information when available20–22 (e.g. by using physicochemical properties of protein cavities).
PCM has been applied to a number of target families including glucocorticoid receptors,23 amine G-protein coupled receptors (GPCR),24–26 melanocortin receptors,27 a wide range of kinases,18,28–32 HIV protease and dengue virus NS3 proteases.33–37 These models successfully identified the discriminating residues for DNA binding,23 mutations contributing to HIV resistance38 and a correlation between the peptide substrate and the protease kinetics.39 For further information the reader is referred to three recent PCM reviews.22,40,41 To date, PCM has been applied on various enzyme families including trypsin-like serine proteases and other members of the protease family (aspartic, cysteine and metallo proteases).42,43 In this study we focused exclusively on serine proteases and addressed their selectivity towards inhibitors.
Here, our main focus is to model the inhibition of serine proteases by small molecules. Firstly, we generate a PCM model to predict the affinity of small molecules for the serine protease family. Secondly, we benchmark 21 protein descriptors, comprising both whole sequence and amino acid descriptors. Finally, we also aim to identify the structural features of both compounds and targets affecting compound affinity toward thrombin (THRB), trypsin (TRY) and coagulation factor 10 (FA10).
Materials and methods
Dataset
The dataset used to generate the models comprised 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 625 distinct inhibitors (20
625 distinct inhibitors (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 213 data points) assayed against 67 protein targets (20
213 data points) assayed against 67 protein targets (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 213 datapoints, matrix ∼30% complete). The complete dataset was obtained from the ‘Directory of Useful Decoys-Enhanced’ (DUD-E),44 Binding Database (Binding DB),45 ZINC46 and ChEMBL-17 databases (10.6019/CHEMBL.database.17).10,47–49 The number of known compound activities per target is given in ST1 (ESI†) along with their UniProt and PDB IDs.50 pChEMBL (−log
213 datapoints, matrix ∼30% complete). The complete dataset was obtained from the ‘Directory of Useful Decoys-Enhanced’ (DUD-E),44 Binding Database (Binding DB),45 ZINC46 and ChEMBL-17 databases (10.6019/CHEMBL.database.17).10,47–49 The number of known compound activities per target is given in ST1 (ESI†) along with their UniProt and PDB IDs.50 pChEMBL (−log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10(activity (nM))) values of five types of activity values were used (IC50, Ki, AC50, EC50 and Kd). The pChEMBL value (Fig. SF1, ESI†) ranges from 3.4 to 11.7. According to this distribution, the dataset contains 15
10(activity (nM))) values of five types of activity values were used (IC50, Ki, AC50, EC50 and Kd). The pChEMBL value (Fig. SF1, ESI†) ranges from 3.4 to 11.7. According to this distribution, the dataset contains 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 375 active datapoints and 4821 inactive datapoints.
375 active datapoints and 4821 inactive datapoints.
Assay identity descriptor (AID)
Publicly available IC50, AC50 and EC50 types are heterogeneous and assay specific.12,51 Thus, in order to combine datapoints with different bioactivity values, we added a binary identifier to the compound and target descriptors, which specifies the activity type (IC50, Ki, AC50, EC50 or Kd) corresponding to each datapoint. We term these descriptors as assay identity descriptors (AID). Formally, AID is defined as:AID(i,j) = δ(i − j)(i ∈ 1, ![[left over right harpoons]](https://www.rsc.org/images/entities/char_21cb.gif) , Ndatapoints, j ∈ 1, , Ndatapoints, j ∈ 1, ![[left over right harpoons]](https://www.rsc.org/images/entities/char_21cb.gif) , NAT) , NAT) |
Chemical descriptors
Molecules were standardized by applying the following filters: “Remove Fragments”, “Neutralize”, “Remove explicit hydrogens”, “Clean 2D”, “Clean 3D” and “Tautomerize” using JChem standardizer, JChem 6.3.1, 2013, ChemAxon (http://www.chemaxon.com). Subsequently, 188 MOE physicochemical descriptors and 256 bit circular fingerprints (radius = 2) were calculated using MOE 2012.1052 and RDKit (http://www.rdkit.org) respectively.53Protein descriptors
Sequences were aligned using the Blosum62 matrix in Clustalw,54,55 whereas the superimposition and structural alignment of drug targets was performed using Chimera 1.6.56 Binding site residues were selected on the basis of a 3D structural alignment of the targets. As structural information was not available for some of the proteins, sequence alignment of the cavities in homologous proteins was used instead. Six amino acid (alignment dependent) descriptors, namely Z-scales, FASGAI, MS-WHIM, protein features, ST scales and T scales15,16 were calculated for the aligned binding site residues with the function AA_descs of the R package camb.57 Descriptor values for sequence alignment gaps were set to zero. The following full protein sequence descriptors (alignment independent descriptors) were calculated using Protpy:19 amino acid composition (AAC), dipeptide composition (DPC), autocorrelation parameters, Moran autocorrelation (MA), normalised Moreau–Broto autocorrelation (MBA), sequence order coupling number (SOCN), Geary autocorrelation (GA), quasi sequence order (QSO) and composition, transition and distribution (CTD) descriptors. Additionally, the Profeat58 descriptor, which is a combination of all full protein sequence descriptors mentioned above, was computed.In order to investigate the predictive power of 3D target information,59 the volume of the binding pocket of each target was calculated using trj_cavity.60 Trj_cavity is a protein cavity analysis software, designed to characterise the cavities present in protein structures extracted from molecular dynamic (MD) trajectories. By using an optimised MD protocol, it identifies the binding pocket, whose coordinates are given by the user, and calculates the static cavity volume. The cavity volume was employed as a baseline to compare the predictive signal alignment dependent and independent descriptors.
Model training
Parameter optimization
In order to reduce the dimensionality of the input space, the recursive feature elimination (RFE) method was applied on both compound and target descriptors.62,63 Random forest models were tuned using number of trees (ntree) equal to 500 and the default number of variables at each layer (mtry = number of variables/3 in case of regression). Model training was performed using the train function of the R package caret.61Model validation
The partitioning of the data set into a training (70% of the data) and a test set (30% of the data) was performed using the createDataPartition function of the R package caret.64 Nested cross-validation and grid search was used for parameter optimisation by dividing the training set into five folds (k = 5). A model was trained on k − 1 folds, which was used to predict the bioactivities of the remaining fold. This procedure was repeated k times for each combination of parameter values. The combination of parameter values displaying the lowest average root mean square error (RMSE) value along the k folds was considered as optimal. Then, a model was built on the whole training set using these values for the parameters.In order to evaluate the extrapolation capabilities of the model to novel serine proteases, we employed leave-one-target-out (LOTO) validation. In LOTO, all datapoints corresponding to a given target/protein were held out of the training set. Subsequently, a model was trained on the remaining data, and the bioactivities for the hold out set were predicted. This type of analysis helped in assessing how well a PCM model predicts the bioactivities for a set of compounds on a target for which no information was presented to the model during the training phase.65–67
In order to determine the predictive power of the models on the test set, two metrics were used: R2 (coefficient of determination) and the root mean square error (RMSE) of prediction (explained in the ESI†).68 A model is considered as predictive if the R2 value on test set is higher than 0.6 and the RMSE less than 1 log units.68
Applicability domain
The applicability domain (AD) of a model is defined as the extent of chemical space, and target in PCM, to which a model can be reliably applied.69 There are various approaches to determine the AD and these highlight the limitations of the model. For example, k-nearest neighbours, probability density distribution, bounding box, distance measures and kernel methods such as Gaussian processes.70–75 Here we used the k nearest neighbours algorithm to determine the AD of the PCM models in the following way. The distance of each compound to its five nearest neighbours in the training set was calculated. Then, the mean similarity of that compound to its neighbours was plotted against the compound's absolute error in prediction, which is defined as the absolute value of the difference between the predicted and the observed bioactivities. Here, the aim is to look for a similarity threshold above which our PCM model can reliably predict the activity of new compounds. To do that, a criterion was set for our PCM model, which states that a PCM model is considered to be predictive if it has a correlation of 0.6 or above and an RMSE less than 1 log units.Feature analysis
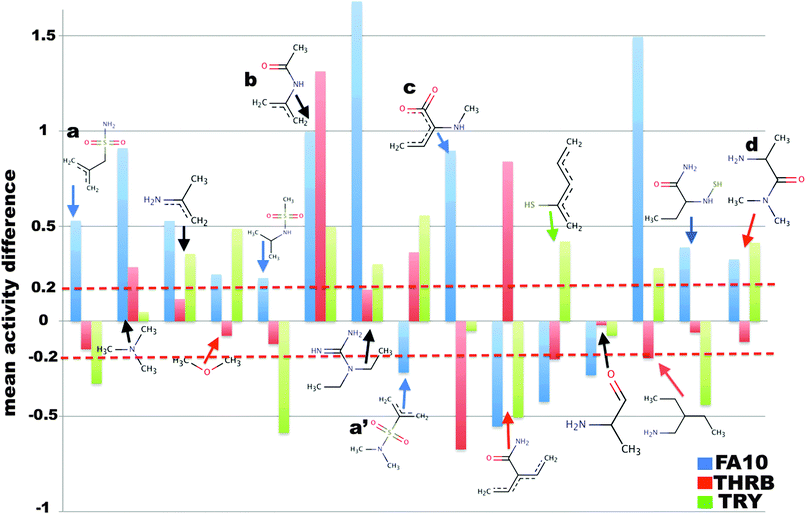
Each bit in the compound descriptors encodes the presence or absence of a chemical substructure, whereas the values of the Z-scales account for physicochemical properties of the amino acids. The influence of each compound substructure and amino acid property on bioactivity was evaluated in the following way. The value of the descriptor was set to zero in all compound or amino acid descriptors presenting it. Then, a PCM model was used to predict the bioactivity of compounds using the updated descriptors.This procedure was repeated for all compound and amino acid descriptors. The analysis was calculated on the basis of the final PCM model trained on all bioactivities, however, due to a lack of data in the training set, chance correlations could happen in the case of certain features of compounds. Here, the threshold (estimated number of datapoints per target) required for a model to reliably predict the feature analysis was not analysed as it is out of the scope of the current study. Instead the average effect of a feature was calculated as the difference in the predicted activity of a compound with and without a given compound substructure or amino acid Z-scale, indicating whether its average influence is beneficial or deleterious on bioactivity as was done previously.67,76 A cut-off value of ±0.2 pChEMBL units equal to the mean of activity difference distribution was established to discriminate which compound substructures or amino acid properties influence the bioactivity (Fig. SF4, ESI†). This analysis was only conducted on THRB, FA10 and TRY, due to the high number of datapoints annotated on these targets, which guarantees statistical robustness.
Results and discussion
Characterization of the biological space
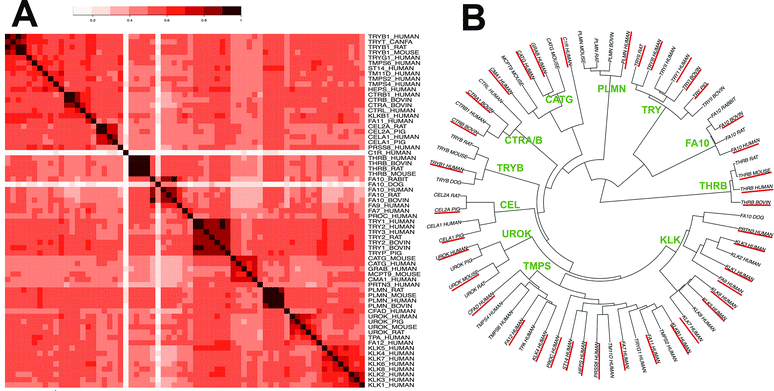
In order to characterise the biological space, we measured the average sequence similarity between all 67 targets using binding site amino acid descriptors. The similarity ranged from 43% to 58%. Table ST2 (ESI†) contains all binding site amino acids considered. The clustering of similar targets in the heat map (Fig. 1A) using binding site residues can also be observed in the clusters of the phylogram (Fig. 1B). In total, we observed 11 big clusters, corresponding to the following sub-families: thrombin (THRB), coagulation factor (FA10), trypsin (TRY), plasminogen (PLMN), cathepsin-G (CATG), chymotrypsin (CTR α/β), tryptase (TRYB), elastase (CEL), urokinase (UROK), transmembrane protease serine (TMPS) and kallikrein (KLK). Homologs of each subfamily were clustered together in the tree, however a few exceptions were observed for members of the following families: FA12, FA11, FA7, serine protease hepsin (HEPS), prostasin (PRSS8) and TMPS2. The targets THRB, TRY and FA10 clustered together suggesting that they are similar enough to enable PCM to inter- and extrapolate compound bioactivity among them. However, there are binding site variations, which might be responsible for the increase or decrease of compound binding affinity across these three proteases.The combined chemical and biological space, termed here as compound–target interaction space, was visualized using both compound and target descriptors. A PCA analysis (Fig. 2) showed that each compound–target group is separated from the others due to ∼41% sequence divergence and the structural diversity of the compounds. However, the three biggest clusters of compound–target pairs in the figure (green: FA10, purple: THRB, pink: TRY) lie close to each other in PCA space enabling the discovery of selective features of these targets. The rest of the targets were found as an overlapping cluster on the top left corner of the PCA plot, explaining the widened chemical and biological space by addition of combined bioactivity data. When the loadings of each principal component were investigated, it was found that maximum variance was explained by the binding pocket sequence descriptors in the biological space. PC1 is loaded mainly by first and second principal components of the Z-scales (34% variance), which summarises the lipophilicity (Z1) and steric bulk properties (Z2) of amino acids, whereas, PC2 (26% variance) is more related to Z-3 of the most varied amino acids in the pocket sequence (e.g. position 56, 140, 177 and 231), explaining the properties like electrophilicity and electronegativity of amino acids.77 PCA also showed that the orthologs are within the targets space defined by FA10, THRB, and TRY. Hence we expected to be able to include them in our data set.
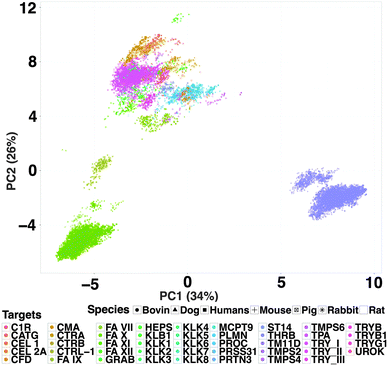 | ||
| Fig. 2 Chemical and biological space (compound–target interaction space) of protease inhibitor dataset. The PCA analysis of chemical descriptors and amino acid descriptors shows an overlap of compound properties per target in PC1 and PC2 space. PC1 (34% variance) shows that the target descriptors influence the spread of data points, clustering the compounds belonging to FA10 and TRY closer to each other, with THRB forming an outer cluster (also shown in phylogram Fig. 1B). PC2 explains the distinct clusters of seven different species (represented with different shapes) used in the study. The dataset has maximum information of human targets, however, the additional orthologous information is distinctly visible using binding pocket features. | ||
PCM model
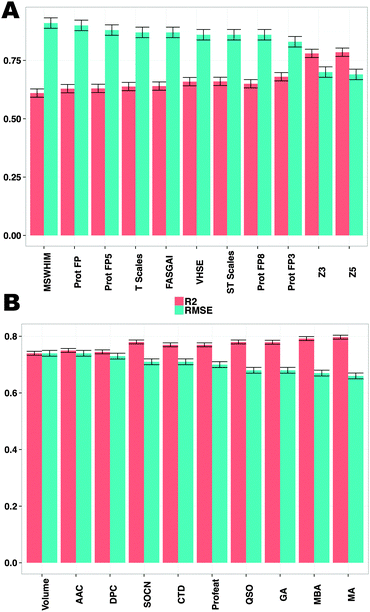
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 213 data points on 67 protease targets were employed in a PCM model using structural fingerprints and protein descriptors. All protein descriptors showed an approximately similar performance. However, the Z-scales combined good performance and interpretability and were used in the final model. The feature type of each descriptor is added for clarity to the reader, FSD indicates the “Full Sequence Descriptor” whereas BSD indicates “Binding Site Descriptor”
213 data points on 67 protease targets were employed in a PCM model using structural fingerprints and protein descriptors. All protein descriptors showed an approximately similar performance. However, the Z-scales combined good performance and interpretability and were used in the final model. The feature type of each descriptor is added for clarity to the reader, FSD indicates the “Full Sequence Descriptor” whereas BSD indicates “Binding Site Descriptor”
| Features | Feature type | R test 2 | RMSEtest |
|---|---|---|---|
| Moran autocorrelation (MA) | FSD | 0.78 | 0.66 |
| Moreau–Broto autocorrelation (MBA) | FSD | 0.78 | 0.67 |
| Z-scales (3) | BSD | 0.78 | 0.70 |
| Quasi-sequence order (QSO) | FSD | 0.78 | 0.68 |
| Geary autocorrelation (GA) | FSD | 0.78 | 0.68 |
| Z-scales (5) | BSD | 0.78 | 0.69 |
| Sequence order coupling numbers (SOCN) | FSD | 0.78 | 0.71 |
| Composition, transition and distribution (CTD) | FSD | 0.77 | 0.70 |
| ProFeat | FSD | 0.77 | 0.70 |
| Amino acid composition (AAC) | FSD | 0.75 | 0.74 |
| Dipeptide composition (DPC) | FSD | 0.74 | 0.73 |
| Volume | BSD | 0.74 | 0.74 |
| ProtFP PCA (3) | BSD | 0.68 | 0.83 |
| ST-scales | BSD | 0.66 | 0.86 |
| VHSE | BSD | 0.66 | 0.86 |
| ProtFP PCA (8) | BSD | 0.65 | 0.86 |
| FASGAI | BSD | 0.64 | 0.87 |
| T-scales | BSD | 0.64 | 0.87 |
| ProtFP PCA (5) | BSD | 0.63 | 0.88 |
| ProtFP (feature) | BSD | 0.63 | 0.91 |
| MSWHIM | BSD | 0.61 | 0.91 |
| Methods | Descriptors | R test 2 | RMSEtest |
|---|---|---|---|
| a Mean value of correlation coefficient and predicted errors (RMSE). | |||
| QSAR (validated per target) | Circular FP | 0.35 ± 0.27a | 1.05 ± 0.27a |
| QSAR (global) | Circular FP | 0.38 | 1.09 |
| PCM (validated per target) | Circular FP, Z3 | 0.64 ± 0.23a | 0.66 ± 0.20a |
| PCM (global) | Circular FP, Z3 | 0.78 | 0.70 |
Although, the predictive power of MBA and SOCN full sequence descriptors (Table 1) was comparable with binding pocket descriptors (mean Rtest2 = 0.78 ± 0.01, mean RMSEtest = 0.68 ± 0.03 log units) (Fig. 3), full sequence descriptors do not permit a biologically meaningful interpretation of the models; hence, 3 Z-scales were used for generation of the final PCM model.
The high performance of models trained on binding site volume indicates that descriptors accounting for structural information of the binding site lead to models displaying slightly worse performance than those trained on more sophisticated protein information as described above. Thus, we anticipate that using more complex structural cavity descriptors alone, or in combination with amino acid or full protein sequence descriptors, is likely to increase model performance.
Comparison of PCM with QSAR
In order to assess whether the inclusion of explicit protein information improves model performance, we trained models on exclusively compound descriptors using the datapoints annotated on a given target (individual QSAR models), or all available datapoints, termed as global or Family QSAR (Table 2).17 The global QSAR model exhibited significantly worse performance than PCM (RMSEQSAR = 1.09, RMSEPCM = 0.70 pChEMBL units). Similarly, PCM also outperformed per target QSAR models, with mean RMSEQSAR and RMSEPCM values on the test set of 1.05 ± 0.27, and 0.66 ± 0.20 pChEMBL units, respectively. This indicates that the explicit inclusion of target information in PCM improves the prediction of bioactivities of the compounds on this dataset.Leave one target out (LOTO) validation
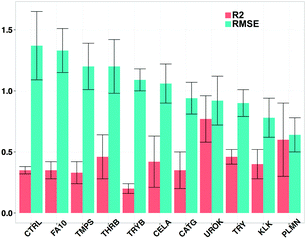
Subsequently, the 67 proteases were grouped into 11 sub-families according to their similarity, which was calculated on binding site amino acid descriptors (Fig. 1B). These groups contained both orthologs and paralogs when applicable. Fig. 4 and Table 3 report the sub-family averaged leave one target out (LOTO) performance. Additionally, the individual performance of LOTO validation on each target is shown in ST3 in the ESI.†| Target sub family | Mean Rtest2 | Mean RMSEtest (log units) |
|---|---|---|
| CTRL | 0.35 ± 0.07 | 1.37 ± 0.57 |
| FA10 | 0.35 ± 0.22 | 1.33 ± 0.55 |
| TMPS | 0.33 ± 0.29 | 1.29 ± 0.64 |
| THRB | 0.46 ± 0.37 | 1.20 ± 0.45 |
| TRYB | 0.20 ± 0.09 | 1.09 ± 0.19 |
| CELA | 0.42 ± 0.43 | 1.06 ± 0.33 |
| CATG | 0.36 ± 0.37 | 0.94 ± 0.31 |
| UROK | 0.77 ± 0.38 | 0.92 ± 0.41 |
| TRY | 0.46 ± 0.17 | 0.90 ± 0.29 |
| KLK | 0.40 ± 0.32 | 0.78 ± 0.52 |
| PLMN | 0.60 ± 0.29 | 0.64 ± 0.28 |
LOTO validation was performed to assess the extrapolation ability of PCM on the target space. We obtained a mean predictive performance (R2) of 0.42 ± 0.15 for all 67 targets with a prediction error (RMSE) of 1.03 ± 0.22 pChEMBL units. LOTO models for CTRL, FA10, TMPS and THRB displayed poor performance with mean R2/RMSE values of (0.35 ± 0.07/1.37 ± 0.57, 0.35 ± 0.22/1.33 ± 0.55, 0.33 ± 0.29/1.29 ± 0.64 and 0.46 ± 0.37/1.20 ± 0.45 respectively). This decrease in performance was expected, as 35% of the datapoints in the dataset are annotated on FA10 and THRB. Hence, removing either of them removes a large fraction of the compound–target interaction space and thus deteriorates model performance. On the other extreme, TMPS and CTRL are annotated with 0.85% of the datapoints and present an average similarity to the other targets considered of ∼43% (Fig. 1B), which is plausibly the reason for the poor performance of their corresponding LOTO models. The low performance of the LOTO model for the CTRL subfamily, with an RMSE equal to 1.37 ± 0.57, likely arises from the low similarity of the members of this family with respect to the other proteases considered here. This can be seen in Fig. 1A, where the CATG subfamily clusters along with HEPS, KLKB1 and other diverse member of TMPS. Taken together, these data indicate that the performance of LOTO models is correlated to the presence of similar proteases in the training set and wherever a diverse protein target is present with less information, the predictability of PCM models decreases with an increase in prediction error.
Although all targets show a close resemblance of the binding pocket in the structural alignment (Table ST1, ESI†), there is considerable variation at several positions, namely: 35, 37, 40, 60, 96, 97, 124, 148, 174, 175, 205, 206, 207, 209, 222, and 227. Out of these variations, amino acids at the 35, 39, 60, 93, 140, and 207 positions were found to affect the binding affinity (Model interpretation section). Given the lower similarity between the targets in the training set (average sequence similarity = ∼41%) as compared to previous studies where the similarity threshold was above 90%,34,79 both the interpolation and extrapolation power of PCM on this data set are notable. Thus, PCM appears as a suitable approach to model compound bioactivities on the serine protease family.
Overall, these data suggest that our PCM models display fair extrapolation performance for proteases similar to those present in the training set.
Applicability domain
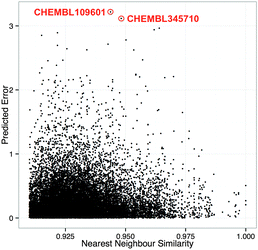
The results from our applicability domain analysis are shown in Fig. 5, which reports the compound similarity averaged over the 5 nearest neighbours against the RMSE values for the compounds in the test set. We obtained high RMSE values, up to 3 pChEMBL units, for compounds displaying a neighbour-averaged similarity value below 0.92. RMSE values gradually decrease, to a minimum value of 0.02 pChEMBL units, as the compound neighbour-averaged similarity increases. In practice, a new compound exhibiting a neighbour-averaged similarity value equal to or greater than 0.90 is likely to be predicted with good accuracy. Nevertheless, we observed some outliers. For instance, two compounds (CHEMBL345710, CHEMBL109601) having a similarity value of ∼0.94 were found to have an exceptionally high prediction error, namely 3.21 and 3.11 pChEMBL units on FA10 and CATG targets. One of these compounds (CHEMBL345710) displays pChEMBL values of 4.20, 4.67 and 6.07 towards FA10, THRB and TRY, whereas the other compound (CHEMBL109601) exhibits bioactivity values of 4.08, 6.27, 8.07 and 6.36 pChEMBL units towards CATG, TRY, CEL and CMA. Together, these observations indicate that our models could not predict the selectivity of these two compounds towards FA10 and CATG, as the bioactivities of their closest neighbours are considerably higher, namely more than 2 pChEMBL units.In conclusion, these data indicate that estimating the error for individual predictions constitutes a valuable source of information in PCM-guided drug discovery campaigns.80
We also performed an applicability domain analysis on the target space. However, we did not observe a correlation between target similarity and error in prediction, likely due to the fact that most of the proteases are similar to each other (average sequence similarity = 46%; Fig. SF2, ESI†). However, the addition of diverse target space (protein sequences of KLK, TMPS, HEPS) is also one of the limitations of poor predictability of the model. There were not enough datapoints available against these targets in the training set; also the compound space was equally sparse, which resulted in poor prediction performance.
Chemical interpretation of the models
This analysis was performed on three targets, namely THRB, TRY and FA10. Fig. 6 reports the influence of compound substructures and binding site amino acid properties on bioactivity. A tertiary sulphonamide (Fig. 6 substructure: a′) was singled out to be important for selectivity against FA10. Absence of this substructure decreased the predicted activity of the compound on average by 0.27 ± 0.65 pChEMBL units against FA10, whereas it increased the bioactivity against THRB and TRY by on average 0.36 ± 0.60 and 0.55 ± 0.73 pChEMBL units, respectively. Thus, the presence of tertiary sulphonamides appears correlated to compound affinity towards FA10 and uncorrelated to affinity on THRB and TRY. We validated this prediction using the 3D crystallographic structures of FA10 and THRB. These structures let us confirm that a tertiary sulphonamide is implicated in strong hydrogen bonds with the backbone residue GLY-219 of FA10 (Fig. 7A). The other important residues of the P1-motif of proteases,81 namely 99, 215 and 219 appear to be involved in ligand binding through van der Waals and electrostatic interactions. However, no significant interaction was observed between this compound substructure and the binding pocket of THRB. This structural analysis confirms the prediction that tertiary sulphonamides are specific for FA10 activity (Fig. 7A).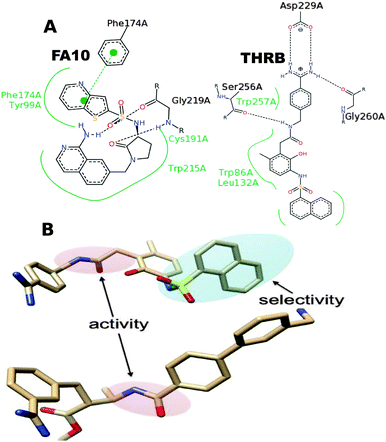 | ||
| Fig. 7 Ligand interaction of compounds containing sulphonamide feature with binding pockets of FA10 (1F0R)82 and THRB (2BDY).83 (A) The selective behaviour of the sulphonamide feature (Fig. 6a′) already predicted by our PCM model is quite evident in this part of the figure. The interactions between the ligands and targets in the crystal structure show the involvement of sulphonamide with binding pocket residues of FA10 protein (GLY-219), however, no interaction is observed in the THRB protein. Interaction figures are generated using Poseview.84 (B) The two features responsible for activity (secondary amide) and selectivity (tertiary sulphonamide) are shown in THRB and FA10 inhibitors. The sub-structural features of the compounds identified by the protease PCM model were validated by analysing the interactions in crystallised protein structures and visualised in (A) and (B) of this figure. | ||
Similarly, absence of a primary sulphonamide (Fig. 6 substructure: a) was observed to increase the predicted activity of the compound on this target. Comparing the binding modes of compounds containing tertiary sulphonamides (Fig. 6a′) and primary sulphonamides (Fig. 6a) features led us to another interesting investigation. The crystallographic structures of both complexes were superimposed and showed that the binding mode of the ligands containing tertiary sulphonamides is different from that of ligands containing primary sulphonamide, and this difference leads to fewer interactions. This structural analysis confirms the validity of the model interpretation pipeline proposed here.
Furthermore, we identified that a secondary amide (Fig. 6b) on average contributes to interactions (hydrogen bonding) with the backbone residues SER-256 of the THRB receptor (2BDY, 1EZQ, 1FXY). The feature analysis for this particular compound substructure however suggested that its absence could increase the predicted bioactivity of the compound on these three targets.
Absence of methylamino-2-butenoic acid was predicted to be beneficial for compound activity on average against FA10 but detrimental against THRB and TRY (Fig. 6c). However, only one compound in our dataset exhibited this feature, and we could not find structural evidence in the literature for the interaction of this feature with THRB, TRY or FA10. Thus, the paucity of experimental data does not permit a complete interpretation of the general influence of this feature on the inhibition of these three proteases.
In addition to these features, we predicted that absence of a prolinamide decreased the predicted bioactivity against THRB by on average 0.10 ± 0.66 pChEMBL units (Fig. 6d), whereas the inverse picture was predicted for FA10 and TRY. When investigating the crystallographic structures available for THRB (PDB IDs: 1AE8 and 3RMM), we found that most of the THRB inhibitors containing this feature interact with GLY-216 and GLY-219. However, no crystal evidence was found for TRY and FA10. Hence, we anticipate that a prolinamide might be a selective feature for THRB and is beneficial for activity against THRB.
Biological interpretation of the models
Next, we analysed the importance of binding site amino acid properties on compound bioactivity (Fig. SF3, ESI†). Residues at positions 35, 39, 60, 93, 140, and 207 were predicted to affect compound bioactivity on average by more than 0.2 pChEMBL units. The presence of an arginine and leucine in THRB at positions 35, 39 and 60 was predicted to be beneficial for bioactivity. However, asparagine, phenylalanine and histidine in FA10, and serine, phenylalanine and lysine in TRY at the same positions, were predicted to decrease compound bioactivity by on average 0.4 ± 0.01 pChEMBL units. This effect illustrates the relevance of small and charged residues in the binding site for compound bioactivity, in opposition to large aromatic residues at positions 25, 39 and 60.The absence of a positively charged arginine at position 93 was predicted to be less important for bioactivity in THRB than a negatively charged glutamate in FA10. However, the absence of any positively charged amino acids (arginine/lysine) in the binding site of TRY leads to an average decrease in activity of 0.07 ± 0.36 pChEMBL units. Furthermore, the presence of lysine and tyrosine at positions 140 and 207, respectively, were predicted to increase the bioactivity in FA10 by on average 0.07 ± 1.06 and 0.05 ± 1.06 pChEMBL units respectively. However, as the impact of these residues on bioactivity towards FA10, THRB and TRY is even less than the chosen cut-off (±0.2), no clear conclusion can be made. The positively charged amino acid residues could favour compound–target interactions and the presence of polar residues in the binding pocket of FA10 could be beneficial for compound affinity, however, stronger evidence in the form of interaction fingerprints in addition to sequence descriptors may help to strengthen these claims.
Conclusions
In the present study, we have introduced PCM for the prediction of the potency of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 625 distinct protease inhibitors on a panel of 67 mammalian serine proteases. We have shown that the inclusion of explicit target information improves the prediction of compound bioactivity on serine proteases, as PCM models outperformed both individual QSAR models and a model trained on exclusively compound descriptors using all datapoints (Family QSAR). We have benchmarked the predictive power of a total of 21 protein descriptors, including binding site amino acid and full protein sequence descriptors, as well as 1D protein cavity descriptors, such as cavity volume, and amino acid composition descriptors. We conclude that the binding site amino acid and full protein sequence descriptors provide comparable predictive signal. However, the usage of binding site amino acid descriptors enabled a biologically meaningful interpretation of the models in agreement with the scientific literature.
625 distinct protease inhibitors on a panel of 67 mammalian serine proteases. We have shown that the inclusion of explicit target information improves the prediction of compound bioactivity on serine proteases, as PCM models outperformed both individual QSAR models and a model trained on exclusively compound descriptors using all datapoints (Family QSAR). We have benchmarked the predictive power of a total of 21 protein descriptors, including binding site amino acid and full protein sequence descriptors, as well as 1D protein cavity descriptors, such as cavity volume, and amino acid composition descriptors. We conclude that the binding site amino acid and full protein sequence descriptors provide comparable predictive signal. However, the usage of binding site amino acid descriptors enabled a biologically meaningful interpretation of the models in agreement with the scientific literature.
Similarly, the description of compounds with keyed fingerprints has permitted a chemically meaningful interpretation of the PCM models. This analysis has singled out compound substructures influencing compound potency and selectivity towards particular proteases, such as primary and tertiary sulphonamides for the selective inhibition of FA10, methylamino-2-butenoic acid for the inhibition of THRB and TRY, and prolinamide for the selective inhibition of THRB.
Overall, the proteochemometric approaches applied on this dataset of serine proteases enabled us to interpret the target information in a meaningful way, which also shows the benefits/strength of incorporating protein-related information in computational chemogenomics.
Acknowledgements
The authors thank Chad H. G. Allen and Lewis Mervin for proof reading the manuscript. Q.U.A. thanks the Islamic Development Bank and Cambridge Commonwealth Trust for Funding. O.M.L. is grateful to CONACyT (No. 217442/312933) and the Cambridge Overseas Trust for funding. G.v.W thanks EMBL (EIPOD) and Marie Curie (COFUND) for funding. A.B. thanks Unilever and the ERC (Starting Grant ERC-2013-StG 336159 MIXTURE) for funding. ICC thanks the Institut Pasteur and the Pasteur-Paris International PhD programme for funding. TM thanks the Institut Pasteur for funding.Notes and references
- A. P. Russ and S. Lampel, Drug Discovery Today, 2005, 10, 1607–1610 CrossRef.
- G. V Paolini, R. H. B. Shapland, W. P. van Hoorn, J. S. Mason and A. L. Hopkins, Nat. Biotechnol., 2006, 24, 805–815 CrossRef PubMed.
- D. Rognan, Br. J. Pharmacol., 2007, 152, 38–52 CrossRef CAS PubMed.
- J. Xu and A. Hagler, Molecules, 2002, 7, 566–600 CrossRef CAS PubMed.
- S. Ekins, J. Mestres and B. Testa, Br. J. Pharmacol., 2007, 152, 9–20 CrossRef CAS PubMed.
- M. Bieler and H. Koeppen, Drug Dev. Res., 2012, 73, 357–364 CrossRef CAS.
- E. Lounkine, M. J. Keiser, S. Whitebread, D. Mikhailov, J. Hamon, J. L. Jenkins, P. Lavan, E. Weber, A. K. Doak, S. Côté, B. K. Shoichet and L. Urban, Nature, 2012, 486, 361–367 CAS.
- Computational Approaches in Cheminformatics and Bioinformatics, ed. R. Guha and A. Bender, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2011 Search PubMed.
- X. Jalencas and J. Mestres, MedChemComm, 2013, 4, 80 RSC.
- A. Gaulton, L. J. Bellis, A. P. Bento, J. Chambers, M. Davies, A. Hersey, Y. Light, S. McGlinchey, D. Michalovich, B. Al-Lazikani and J. P. Overington, Nucleic Acids Res., 2012, 40, D1100–D1107 CrossRef CAS PubMed.
- C. Kramer and R. Lewis, Curr. Top. Med. Chem., 2012, 12, 1896–1902 CrossRef CAS.
- T. Kalliokoski, C. Kramer, A. Vulpetti and P. Gedeck, PLoS One, 2013, 8, e61007 CAS.
- H. Kubinyi, in Chemogenomics in Drug Discovery, ed. H. Kubinyi and G. Müller, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, FRG, 2004 Search PubMed.
- P. Willett, Annu. Rev. Inf. Sci. Technol., 2009, 43, 1–117 CrossRef.
- G. J. van Westen, R. F. Swier, J. K. Wegner, A. P. Ijzerman, H. W. van Vlijmen and A. Bender, J. Cheminf., 2013, 5, 41 CAS.
- G. J. van Westen, R. F. Swier, I. Cortes-Ciriano, J. K. Wegner, J. P. Overington, A. P. Ijzerman, H. W. van Vlijmen and A. Bender, J. Cheminf., 2013, 5, 42 CAS.
- J. B. Brown, Y. Okuno, G. Marcou, A. Varnek and D. Horvath, J. Comput.-Aided Mol. Des., 2014, 28, 597–618 CrossRef CAS PubMed.
- D.-S. Cao, G.-H. Zhou, S. Liu, L.-X. Zhang, Q.-S. Xu, M. He and Y.-Z. Liang, Anal. Chim. Acta, 2013, 792, 10–18 CrossRef CAS PubMed.
- D.-S. Cao, Q.-S. Xu and Y.-Z. Liang, Bioinformatics, 2013, 29, 960–962 CrossRef CAS PubMed.
- J. Gao, Q. Huang, D. Wu, Q. Zhang, Y. Zhang, T. Chen, Q. Liu, R. Zhu, Z. Cao and Y. He, Gene, 2013, 518, 124–131 CrossRef CAS PubMed.
- N. Weill, Curr. Top. Med. Chem., 2011, 11, 1944–1955 CrossRef CAS.
- C. R. Andersson, M. G. Gustafsson and H. Strömbergsson, Curr. Top. Med. Chem., 2011, 11, 1978–1993 CrossRef CAS.
- J. Zilliacus, A. P. Wright, U. Norinder, J. A. Gustafsson and J. Carlstedt-Duke, J. Biochem., 1992, 267, 24941–24947 CAS.
- M. Lapinsh, P. Prusis, T. Lundstedt and J. E. S. Wikberg, Mol. Pharmacol., 2002, 61, 1465–1475 CrossRef CAS.
- T. M. Frimurer, T. Ulven, C. E. Elling, L.-O. Gerlach, E. Kostenis and T. Högberg, Bioorg. Med. Chem. Lett., 2005, 15, 3707–3712 CrossRef CAS PubMed.
- L. Jacob, B. Hoffmann, V. Stoven and J.-P. Vert, BMC Bioinf., 2008, 9, 363–379 CrossRef PubMed.
- M. Lapinsh, S. Veiksina, S. Uhlén, R. Petrovska, I. Mutule, F. Mutulis, S. Yahorava, P. Prusis and J. E. S. Wikberg, Mol. Pharmacol., 2005, 67, 50–59 CrossRef CAS PubMed.
- M. Lapins and J. E. S. Wikberg, BMC Bioinf., 2010, 11 DOI:10.1186/1471-2105-11-339.
- M. W. Karaman, S. Herrgard, D. K. Treiber, P. Gallant, C. E. Atteridge, B. T. Campbell, K. W. Chan, P. Ciceri, M. I. Davis, P. T. Edeen, R. Faraoni, M. Floyd, J. P. Hunt, D. J. Lockhart, Z. V. Milanov, M. J. Morrison, G. Pallares, H. K. Patel, S. Pritchard, L. M. Wodicka and P. P. Zarrinkar, Nat. Biotechnol., 2008, 26, 127–132 CrossRef CAS PubMed.
- V. Subramanian, P. Prusis, L.-O. Pietilä, H. Xhaard and G. Wohlfahrt, J. Chem. Inf. Model., 2013, 53, 3021–3030 CrossRef CAS PubMed.
- M. I. Davis, J. P. Hunt, S. Herrgard, P. Ciceri, L. M. Wodicka, G. Pallares, M. Hocker, D. K. Treiber and P. P. Zarrinkar, Nat. Biotechnol., 2011, 29, 1046–1051 CrossRef CAS PubMed.
- G. Subramanian and M. Sud, ACS Med. Chem. Lett., 2010, 1, 395–399 CrossRef CAS.
- M. Junaid, M. Lapins, M. Eklund, O. Spjuth and J. E. S. Wikberg, PLoS One, 2010, 5, e14353 CAS.
- G. J. P. van Westen, A. Hendriks, J. K. Wegner, A. P. Ijzerman, H. W. T. van Vlijmen and A. Bender, PLoS Comput. Biol., 2013, 9, e1002899 CAS.
- K. M. Doherty, P. Nakka, B. M. King, S.-Y. Rhee, S. P. Holmes, R. W. Shafer and M. L. Radhakrishnan, BMC Bioinf., 2011, 12, 477–496 CrossRef PubMed.
- A. Kontijevskis, P. Prusis, R. Petrovska, S. Yahorava, F. Mutulis, I. Mutule, J. Komorowski and J. E. S. Wikberg, PLoS Comput. Biol., 2007, 3, e0424 Search PubMed.
- S. Jayaraman and K. Shah, In Silico Biol., 2008, 8, 427–447 CAS.
- M. Lapins, M. Eklund, O. Spjuth, P. Prusis and J. E. S. Wikberg, BMC Bioinf., 2008, 9, 181–192 CrossRef PubMed.
- P. Prusis, M. Lapins, S. Yahorava, R. Petrovska, P. Niyomrattanakit, G. Katzenmeier and J. E. S. Wikberg, Bioorg. Med. Chem., 2008, 16, 9369–9377 CrossRef CAS PubMed.
- G. J. P. van Westen, J. K. Wegner, A. P. IJzerman, H. W. T. van Vlijmen and A. Bender, MedChemComm, 2011, 2, 16–30 RSC.
- I. C. Ciriano, Q. ul Ain, V. Subramanian, E. B. Lenselink, O. M. Lucio, A. P. IJzerman, G. Wohlfahrt, P. Prusis, T. E. Malliavin, G. J. Van Westen and A. Bender, MedChemComm Search PubMed , in revision.
- H. Strömbergsson, A. Kryshtafovych, P. Prusis, K. Fidelis, J. E. S. Wikberg, J. Komorowski and T. R. Hvidsten, Proteins, 2006, 65, 568–579 CrossRef PubMed.
- A. M. Wassermann, H. Geppert and J. Bajorath, J. Chem. Inf. Model., 2009, 49, 2155–2167 CrossRef CAS PubMed.
- M. M. Mysinger, M. Carchia, J. J. Irwin and B. K. Shoichet, J. Med. Chem., 2012, 55, 6582–6594 CrossRef CAS PubMed.
- T. Liu, Y. Lin, X. Wen, R. N. Jorrisen and M. K. Gilson, Nucleic Acids Res., 2007, 35, 198–201 CrossRef PubMed.
- J. J. Irwin, T. Sterling, M. M. Mysinger, E. S. Bolstad and R. G. Coleman, J. Chem. Inf. Model., 2012, 52, 1757–1768 CrossRef CAS PubMed.
- A. P. Bento, A. Gaulton, A. Hersey, L. J. Bellis, J. Chambers, M. Davies, F. A. Krüger, Y. Light, L. Mak, S. McGlinchey, M. Nowotka, G. Papadatos, R. Santos and J. P. Overington, Nucleic Acids Res., 2014, 42, D1083–D1090 CrossRef CAS PubMed.
- S. Jupp, J. Malone, J. Bolleman, M. Brandizi, M. Davies, L. Garcia, A. Gaulton, S. Gehant, C. Laibe, N. Redaschi, S. M. Wimalaratne, M. Martin, N. Le Novère, H. Parkinson, E. Birney and A. M. Jenkinson, Bioinformatics, 2014, 30, 1338–1339 CrossRef CAS PubMed.
- R. Ochoa, M. Davies, G. Papadatos, F. Atkinson and J. P. Overington, Bioinformatics, 2014, 30, 298–300 CrossRef CAS PubMed.
- R. Giegé, FEBS J., 2013, 280, 6456–6497 CrossRef PubMed.
- C. Kramer, T. Kalliokoski, P. Gedeck and A. Vulpetti, J. Med. Chem., 2012, 55, 5165–5173 CrossRef CAS PubMed.
- M. O. E. Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2013.
- G. Landrum, 2011.
- M. Goujon, H. McWilliam, W. Li, F. Valentin, S. Squizzato, J. Paern and R. Lopez, Nucleic Acids Res., 2010, 38, W695–W699 CrossRef CAS PubMed.
- M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson and D. G. Higgins, Bioinformatics, 2007, 23, 2947–2948 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- D. S. Murrell, I. Cortés-Ciriano, G. J. P. van Westen, I. P. Stott, A. Bender, T. Malliavin and R. C. Glen, http://github.com/cambDI/camb, 2014.
- Z. R. Li, H. H. Lin, L. Y. Han, L. Jiang, X. Chen and Y. Z. Chen, Nucleic Acids Res., 2006, 34, W32–W37 CrossRef CAS PubMed.
- T. Liu, Y. Lin, X. Wen, R. N. Jorissen and M. K. Gilson, Nucleic Acids Res., 2007, 35, D198–D201 CrossRef CAS PubMed.
- T. Paramo, A. East, D. Garzón, M. B. Ulmschneider and P. J. Bond, J. Chem. Theory Comput., 2014, 10, 2151–2164 CrossRef CAS.
- M. Kuhn, J. Wing, S. Weston, A. Williams, C. Keefer, A. Engelhardt and T. Cooper, 2013.
- M. Kuhn, J. Stat. Softw., 2008, 28, 1–26 Search PubMed.
- X. Lin, F. Yang, L. Zhou, P. Yin, H. Kong, W. Xing, X. Lu, L. Jia, Q. Wang and G. Xu, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2012, 910, 149–155 CrossRef CAS PubMed.
- T. Pahikkala, A. Airola, S. Pietilä, S. Shakyawar, A. Szwajda, J. Tang and T. Aittokallio, Briefings Bioinf., 2014 Search PubMed.
- C. Kramer and P. Gedeck, J. Chem. Inf. Model., 2010, 50, 1961–1969 CrossRef CAS PubMed.
- P. J. Ballester and J. B. O. Mitchell, J. Chem. Inf. Model., 2011, 51, 1739–1741 CrossRef CAS PubMed.
- G. J. P. van Westen, J. K. Wegner, P. Geluykens, L. Kwanten, I. Vereycken, A. Peeters, A. P. Ijzerman, H. W. T. van Vlijmen and A. Bender, PLoS One, 2011, 6, e27518 CAS.
- A. Tropsha and A. Golbraikh, Curr. Pharm. Des., 2007, 13, 3494–3504 CrossRef CAS.
- J. Jaworska, N. Nikolova-jeliazkova and T. Aldenberg, Altern. Lab. Anim., 2005, 33, 445–459 CAS.
- F. Sahigara, D. Ballabio, R. Todeschini and V. Consonni, J. Cheminf., 2013, 5, 27 CAS.
- F. Sahigara, K. Mansouri, D. Ballabio, A. Mauri, V. Consonni and R. Todeschini, Molecules, 2012, 17, 4791–4810 CrossRef CAS PubMed.
- I. Sushko, S. Novotarskyi, R. Körner, A. K. Pandey, V. V. Kovalishyn, V. V. Prokopenko and I. V. Tetko, J. Chemom., 2010, 24, 202–208 CrossRef CAS.
- R. P. Sheridan, J. Chem. Inf. Model., 2013, 53, 2837–2850 CrossRef CAS PubMed.
- R. P. Sheridan, J. Chem. Inf. Model., 2012, 52, 814–823 CrossRef CAS PubMed.
- N. Fechner, A. Jahn, G. Hinselmann and A. Zell, J. Cheminf., 2010, 2, 2 Search PubMed.
- J. Klekota and F. P. Roth, Bioinformatics, 2008, 24, 2518–2525 CrossRef CAS PubMed.
- M. Sandberg, L. Eriksson, J. Jonsson, M. Sjöström and S. Wold, J. Med. Chem., 1998, 41, 2481–2491 CrossRef CAS PubMed.
- A. Bender and R. C. Glen, J. Chem. Inf. Model., 2005, 45, 1369–1375 CrossRef CAS PubMed.
- Q. Huang, H. Jin, Q. Liu, Q. Wu, H. Kang, Z. Cao and R. Zhu, PLoS One, 2012, 7, e41698 CAS.
- I. Cortes-Ciriano, G. J. van Westen, E. B. Lenselink, D. S. Murrell, A. Bender and T. Malliavin, J. Cheminf., 2014, 6, 35 Search PubMed.
- L. Hedstrom, Chem. Rev., 2002, 102, 4501–4524 CrossRef CAS PubMed.
- S. Maignan, J. P. Guilloteau, S. Pouzieux, Y. M. Choi-Sledeski, M. R. Becker, S. I. Klein, W. R. Ewing, H. W. Pauls, A. P. Spada and V. Mikol, J. Med. Chem., 2000, 43, 3226–3232 CrossRef CAS PubMed.
- S. Hanessian, E. Therrien, W. A. L. van Otterlo, M. Bayrakdarian, I. Nilsson, O. Fjellström and Y. Xue, Bioorg. Med. Chem. Lett., 2006, 16, 1032–1036 CrossRef CAS PubMed.
- K. Stierand and M. Rarey, ACS Med. Chem. Lett., 2010, 1, 540–545 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ib00175c |
| This journal is © The Royal Society of Chemistry 2014 |