A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane†
Amelia-Elena
Rotaru‡
*a,
Pravin Malla
Shrestha‡
a,
Fanghua
Liu
ab,
Minita
Shrestha
a,
Devesh
Shrestha
a,
Mallory
Embree
c,
Karsten
Zengler
c,
Colin
Wardman
a,
Kelly P.
Nevin
a and
Derek R.
Lovley
a
aDepartment of Microbiology, University of Massachusetts, Amherst, Massachusetts 01003, USA. E-mail: arotaru@microbio.umass.edu; Fax: +1-413-545-1578; Tel: +1-413-577-3069
bYantai Institute of Coastal Research, Yantai, China
cUniversity of California, San Diego, La Jolla, CA, USA
First published on 18th October 2013
Abstract
Anaerobic conversion of organic wastes and biomass to methane is an important bioenergy strategy, which depends on poorly understood mechanisms of interspecies electron transfer to methanogenic microorganisms. Metatranscriptomic analysis of methanogenic aggregates from a brewery wastewater digester, coupled with fluorescence in situ hybridization with specific 16S rRNA probes, revealed that Methanosaeta species were the most abundant and metabolically active methanogens. Methanogens known to reduce carbon dioxide with H2 or formate as the electron donor were rare. Although Methanosaeta have previously been thought to be restricted to acetate as a substrate for methane production, Methanosaeta in the aggregates had a complete complement of genes for the enzymes necessary for the reduction of carbon to methane, and transcript abundance for these genes was high. Furthermore, Geobacter species, the most abundant bacteria in the aggregates, highly expressed genes for ethanol metabolism and for extracellular electron transfer via electrically conductive pili, suggesting that Geobacter and Methanosaeta species were exchanging electrons via direct interspecies electron transfer (DIET). This possibility was further investigated in defined co-cultures of Geobacter metallireducens and Methanosaeta harundinacea which stoichiometrically converted ethanol to methane. Transcriptomic, radiotracer, and genetic analysis demonstrated that M. harundinacea accepted electrons via DIET for the reduction of carbon dioxide to methane. The discovery that Methanosaeta species, which are abundant in a wide diversity of methanogenic environments, are capable of DIET has important implications not only for the functioning of anaerobic digesters, but also for global methane production.
Broader contextIn this study we report a fundamentally new concept for the microbial ecology of anaerobic digestion, one of the oldest bioenergy strategies. The reliance of methanogenic communities on interspecies electron transfer has been recognized for over forty years, but it has been thought that only H2 or formate served as the interspecies electron carriers. However, the finding that Methanosaeta species can make direct electrical connections with Geobacter species, accepting electrons for the reduction of carbon dioxide to methane, demonstrates that direct interspecies electron transfer (DIET) is an alternative to interspecies H2/formate transfer. DIET appears to predominate over interspecies H2/formate transfer in upflow anaerobic digesters converting brewery waste to methane, and the metatranscriptomic approach described here provides a tool to discriminate between pathways for interspecies electron transfer in other digester designs, treating other types of wastes or biomass. Methanosaeta species are also ubiquitous in methanogenic soils and sediments, suggesting that a substantial portion of global methane production could be derived from DIET. |
Introduction
Anaerobic conversion of organic compounds to methane is one of the few proven, economical, large-scale bioenergy strategies. Methanogenic treatment of wastewaters is already a widespread practice and new approaches to reactor design are expected to further improve this technology and expand its application.1,2Nearly half a century ago, a major breakthrough in the understanding of the function of methanogenic microbial communities was made with the discovery of interspecies H2 transfer.3–5 In interspecies H2 transfer non-methanogenic microorganisms metabolize key fermentation products, such as ethanol and volatile fatty acids, to acetate, which methanogens then convert to methane. This acetate production also releases carbon dioxide and reduces electron carriers in the acetate-producing microbes. The reduced electron carriers are regenerated to the oxidized state via the reduction of protons to H2. Methanogens consume the H2 with the reduction of carbon dioxide to methane. This syntrophic degradation of fermentation intermediates functions well as long as methanogens maintain the concentration of H2 low enough that the production of H2 is thermodynamically favourable. Formate can serve as a substitute for H2 as an interspecies electron carrier.6,7 Interspecies H2/formate transfer has been documented in many defined co-cultures in which H2 and/or formate-donating microorganisms were paired with H2 and/or formate-consuming methanogens.7–10
It has been assumed that the interspecies H2/formate transfer observed in laboratory co-cultures is also the primary mechanism for interspecies electron exchange in anaerobic digesters and other complex methanogenic environments, such as anaerobic soils and sediments. However, it has been difficult to determine the extent of interspecies H2/formate transfer in such environments because of the lack of methods for reliably measuring turnover rates of H2 and formate. A potential alternative to interspecies H2/formate transfer is direct interspecies electron transfer (DIET), in which species exchange electrons through biological electrical connections.11–14
DIET was first documented in co-cultures of Geobacter metallireducens and Geobacter sulfurreducens grown in a medium with ethanol as the electron donor and fumarate as the electron acceptor.11 Interspecies electron exchange was required because G. metallireducens can metabolize ethanol, but cannot use fumarate as an electron acceptor, whereas G. sulfurreducens can reduce fumarate, but cannot metabolize ethanol. Studies with a diversity of mutant strains, deficient in key aspects of interspecies H2/formate transfer or DIET, as well as genome-wide transcriptomic analysis, demonstrated that H2 or formate could not be the interspecies electron carrier.11,12,15 Instead, the co-cultures established electrical connections through the pili of the two Geobacter species, which are electrically conductive.16–18 Consistent with the DIET concept, the Geobacter co-cultures formed large (1–2 mm diameter), electrically conductive aggregates to promote interspecies electron exchange.11
Methanogenic aggregates from a brewery wastewater digester were also electrically conductive, with a temperature dependence characteristic of the metal-like conductivity of Geobacter pili.19Geobacter species were the dominant bacteria, accounting for ca. 25% of the bacterial 16S rRNA gene sequences recovered. A similar abundance of Geobacter species has been observed in many similar brewery waste digesters.20
Methanosaeta species accounted for over 90% of the 16S rRNA sequences recovered that could be attributed to methanogens.19Methanosaeta species can convert acetate to methane but cannot utilize H2 or formate as an electron donor for the reduction of carbon dioxide to methane.21 In accordance with the low abundance of methanogens known to metabolize H2 or formate, the aggregates only slowly converted these compounds to methane.19 Based on these observations it was proposed that DIET, rather than interspecies H2/formate transfer, was the mechanism for interspecies electron exchange within the methanogenic digester aggregates.19 However, this was speculative because it had not been shown that Methanosaeta or any other methanogens were actually capable of accepting electrons via DIET. Furthermore, in order for Methanosaeta species to participate in DIET they would need to reduce carbon dioxide to methane, an unknown metabolic capability in these organisms.
Recent studies with co-cultures suggested that community gene expression patterns are different during DIET than interspecies H2/formate transfer.12,15 Therefore, in order to overcome the challenges of directly tracking the flow of H2 or electrons between microorganisms in complex communities we used gene expression patterns as a diagnostic tool to elucidate mechanisms of interspecies electron exchange in the digester aggregates.
Results and discussion
Evidence for direct electron transfer in digester aggregates
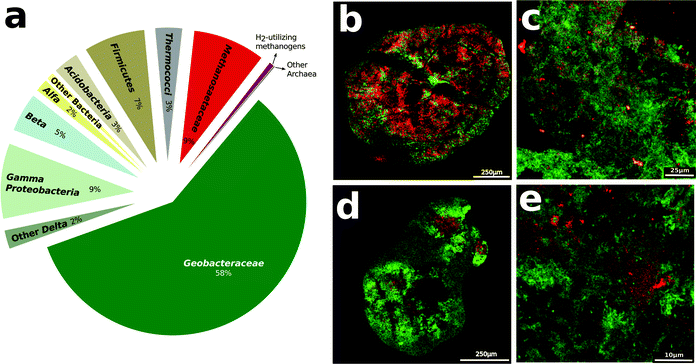
Analysis of gene transcript abundance in the aggregates from the digesters treating simulated brewery wastewater revealed that Methanosaeta species were the predominant and active methanogens (Fig. 1a). Fluorescence in situ hybridization (FISH) confirmed the abundance of Methanosaeta, which were homogenously distributed throughout the aggregates (Fig. 1b–e). Less than 0.6% of gene transcript reads could be ascribed to methanogens capable of metabolizing H2 or formate, and FISH verified that such methanogens were rare (Fig. 1c–e).The metabolism of ethanol, the primary waste in brewery digesters, produces acetate with the release of electrons:
| CH3CH2OH + H2O → CH3COOH + 4H+ + 4e− | (1) |
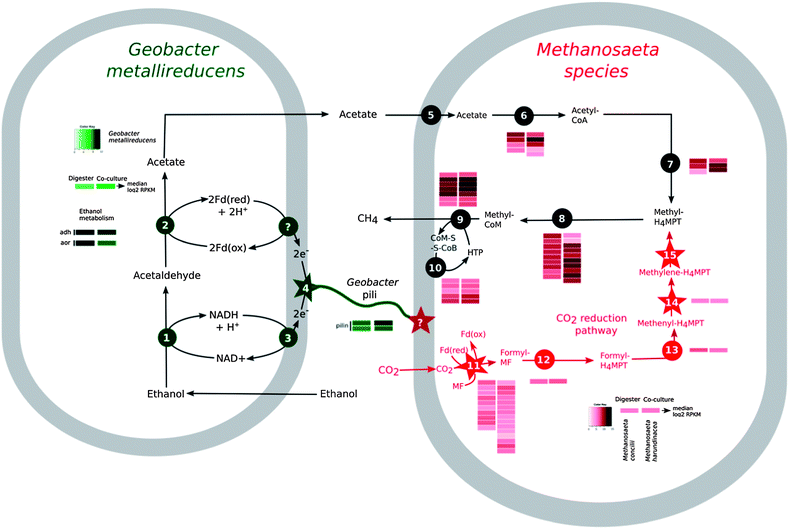
Therefore, it was not surprising that Methanosaeta genes encoding enzymes involved in converting acetate to methane were highly expressed in the reactor aggregates (Fig. 2). However, in order for ethanol to be completely metabolized, the electrons released during ethanol metabolism must be consumed.
In a methanogenic environment the most direct route for consumption of these electrons is the reduction of carbon dioxide to methane. During interspecies H2 transfer the ethanol-metabolizing microorganisms produce H2:
| 2e− + 2H+ → H2 | (2) |
| 4H2 + CO2 → CH4 + 2H2O | (3) |
The inability of Methanosaeta species to use H2, or the H2 substitute formate,21 eliminated the possibility for this mode of electron exchange. However, genes for a complete pathway for carbon dioxide reduction were present and highly expressed in Methanosaeta concilii, the abundant species in the digester aggregates (Fig. 2). This suggested that Methanosaeta species were actively reducing carbon dioxide to methane, possibly with electrons derived by a mechanism other than interspecies H2 or formate transfer.
Previous studies demonstrated that Geobacter species were the most abundant bacteria in the digesters19 and in this study we observed that Geobacter species accounted for most of the gene transcripts recovered, demonstrating their high metabolic activity (Fig. 2, Table S2†). The Geobacter species highly expressed genes for ethanol metabolism (Fig. 2, Table S2†), suggesting that they played an important role in utilizing this primary substrate in the digesters. Geobacter metallireducens is known to transfer electrons derived from ethanol metabolism to Geobacter sulfurreducens via pili11 that are electrically conductive.16,17 The gene for PilA, the structural protein for electrically conductive pili, was highly expressed (Fig. 2, Table S3†), as expected12,15 if the abundant Geobacter species were metabolizing ethanol with direct electron transfer to Methanosaeta.
Direct electron transfer to Methanosaeta in defined co-cultures
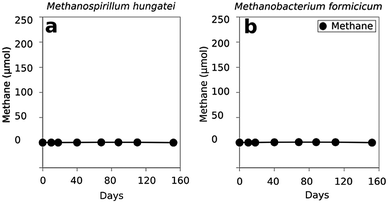
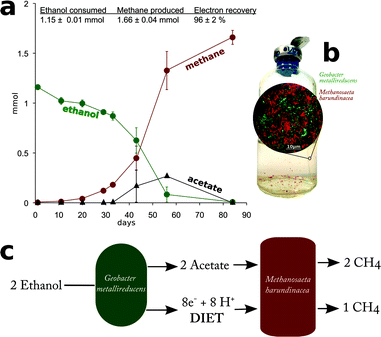
In order to evaluate whether Methanosaeta was capable of functioning in the manner suggested by the metatranscriptomic analysis, Methanosaeta harundinacea, an isolate from another anaerobic digester treating brewery waste22 was co-cultured with Geobacter metallireducens, which served as a representative for the abundant Geobacter species in the digesters. Metabolic modelling has suggested that G. metallireducens is unable to conserve energy to support growth from syntrophic metabolism of ethanol with the production of H2 or formate13,23 and the inability of G. metallireducens to grow via interspecies H2 or formate transfer was further evident from its failure to generate functioning co-cultures with the H2- or formate-utilizing methanogens Methanospirillum hungatei or Methanobacterium formicicum (Fig. 3). These results, coupled with the inability of M. harundinacea to metabolize H2 or formate,22,24 ruled out the possibility of electron exchange via these indirect electron carriers.Yet, G. metallireducens and M. harundinacea grew in co-culture converting ethanol to methane, forming aggregates in which the two species were in close physical proximity (Fig. 4). The amount of methane produced in the co-cultures was consistent with complete conversion of the added ethanol to methane based on the following reactions:
| 2CH3CH2OH + 2H2O → 2CH3COOH + 8H+ + 8e− | (4) |
| 2CH3COOH → 2CH4 + 2CO2 | (5) |
| CO2 + 8e− + 8H+ → CH4 + 2H2O | (6) |
| Sum of reactions: 2CH3CH2OH → 3CH4 + CO2 | (7) |
Each mole of ethanol yielded ca. 1.5 moles of methane (Fig. 4). This indicated that M. harundinacea was not only converting the acetate produced from ethanol to methane (reaction (5)), but was also utilizing the additional electrons available from the conversion of ethanol to acetate (reaction (4)) for methane production (reaction (6)). Metatranscriptomic analysis of the co-culture revealed that the genes for the putative carbon dioxide reduction pathway in M. harundinacea were highly expressed (Fig. 2, Table S3†), as expected if M. harundinacea was directly accepting electrons from G. metallireducens for the reduction of carbon dioxide to methane.
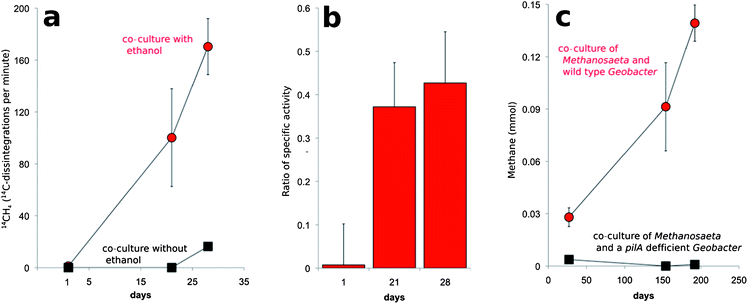
In order to further evaluate this possibility, co-cultures were transferred into fresh medium that was amended with [14C]-bicarbonate. The specific activity (14C disintegrations per minute per mole compound) of the CH4 that was produced was ca. ⅓ of the specific activity measured for CO2 (Fig. 5). This is the result expected according to reactions (4)–(7) in which ⅓ of the methane produced should be derived from CO2 (Fig. 5).
Geobacter species require their electrically conductive pili for extracellular electron transfer to insoluble electron acceptors, such as Fe(III) oxides,16,25 but not for reduction of soluble extracellular molecules that might function as electron shuttles between cells.16Geobacter strains in which the gene for PilA was deleted, were incapable of DIET in G. metallireducens–G. sulfurreducens co-cultures.11,12 In this study, G. metallireducens highly expressed the PilA gene in co-culture with M. harundinacea (Fig. 2, Table S3†), and the PilA-deficient strain of G. metallireducens did not metabolize ethanol or produce methane in co-culture with M. harundinacea (Fig. 5). These results indicate that pili are an important component for electron transfer between G. metallireducens and M. harundinacea.
The stoichiometric conversion of ethanol to methane coupled with the high expression of genes for carbon dioxide reduction in M. harundinacea and the reduction of 14CO2 to 14CH4 at the appropriate specific activity demonstrated that M. harundinacea was capable of accepting electrons from G. metallireducens for the reduction of carbon dioxide to methane. Although genes for carbon dioxide reduction were previously noted in genomes of Methanosaeta species,24 our study is the first to document that Methanosaeta is capable of producing methane from carbon dioxide. The fact that M. harundinacea reduced carbon dioxide under conditions in which interspecies H2/formate transfer was impossible, and that co-cultures could not be established with a strain of G. metallireducens that could not produce conductive pili, indicated that G. metallireducens and M. harundinacea exchanged electrons through a biological electrical connection. This is the first example, of a methanogen participating in DIET.
Methods
Laboratory scale digesters
Microbial aggregates were propagated in the laboratory in three replicate mesophilic (37 °C) laboratory-scale (0.9 liter) digesters.19Strains, media, culturing conditions
Cultures were grown under strict anaerobic conditions in anaerobic pressure tubes or serum bottles sealed with thick butyl rubber stoppers. Geobacter metallireducens, wild type and the PilA-deficient strain, were routinely maintained on Fe(III)-citrate (FC) medium with 10 mM acetate as an electron donor.25 Prior to co-cultivation, all Geobacter strains were adapted to growth on Fe(III) citrate medium with 20 mM ethanol as the substrate for more than three transfers until ethanol metabolism was synchronized in the wild type and the PilA-deficient strain. Methanosaeta harundinacea (JCM-13211) was purchased from the Japanese culture collection and Methanospirillum hungatei (DSM-13809), and Methanobacterium formicicum (DSM-1535) were purchased from the German culture collection DSMZ. The methanogens were grown under the conditions specified by the culture collections.Co-cultures were initiated with 0.5 mL of G. metallireducens, and 1 mL of the methanogen cultures inoculated into 10 mL modified fresh water medium with 20 mM ethanol and carbon dioxide as the only electron acceptor. The medium was modified from a previously described fresh water medium.26 Modifications consisted of boiling the medium to reduce O2 solubility, then cooling under N2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CO2 (80
CO2 (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) for increased gas exchange. Prior to addition of cells the medium was amended from anaerobic sterile stocks with: 20 mM ethanol, vitamins,26 and a premix of 1 mM cysteine, and 0.5 mM Na2S·9H2O. For studies on the stoichiometry of ethanol metabolism co-cultures were grown in 50 mL of medium in 160 mL serum bottles. Samples were withdrawn regularly with N2
20) for increased gas exchange. Prior to addition of cells the medium was amended from anaerobic sterile stocks with: 20 mM ethanol, vitamins,26 and a premix of 1 mM cysteine, and 0.5 mM Na2S·9H2O. For studies on the stoichiometry of ethanol metabolism co-cultures were grown in 50 mL of medium in 160 mL serum bottles. Samples were withdrawn regularly with N2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CO2 (80
CO2 (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) degassed hypodermic syringes to monitor ethanol, acetate, and methane as previously described.15,19
20) degassed hypodermic syringes to monitor ethanol, acetate, and methane as previously described.15,19
For radiotracer experiments, a sterile anaerobic solution of [14C]-bicarbonate (18.7 × 104 Bq per 10 mL) was added to co-cultures to obtain a final concentration of 8.1 × 104 Bq. 14CH4 and 14CO2 were monitored with a gas chromatograph gas proportional counter as previously described.27
Fluorescent in situ hybridization (FISH)
The distribution of microorganisms within thin sections of digester or co-culture aggregates was examined with FISH probes as previously described.28 The probes were: MX825 targeting Methanosaeta,29 MB1174 (ref. 29) targeting Methanobacteriaceae; MS1414 (ref. 29) targeting Methanosarcina; GEO825 (ref. 30) specific for Geobacter species; and GEO1 (ref. 11) specific for Geobacter metallireducens. Samples were imaged with a Leica TCS SP5 microscope as previously described.15mRNA extraction
Digester aggregates were sampled from two independent digesters, and were immediately mixed with RNAlater (Ambion) as described previously.12 Samples of three replicate Geobacter metallireducens and Methanosaeta harudinacea co-cultures were harvested as previously described.12 Samples were then processed immediately or stored at −80 °C.RNA was extracted and mRNA was enriched as described previously.12 The workflow for the metatranscriptomic analysis can be found in Fig. S1.†
Illumina sequencing
Digester mRNA was sequenced with paired end, strand specific RNA sequencing with the dUTP method as previously described23,31 on an Illumina Genome Analyzer II. For the co-cultures, directional libraries were prepared with the ScriptSeq™ v2 RNA-Seq Library Preparation Kit (Epicentre) and single end sequencing was done with a Hi-Seq 2000 following the manufacturer's instructions.Assembly of Illumina reads
All the raw sequencing data were quality checked by visualization of base quality scores and nucleotide distributions. Then the sequences were sorted by trimming of reads and read filtering based on the base quality score and sequence properties such as primer contaminations, N content and GC bias with PRINSEQ.32 We removed sequence reads matching 16S and 23S rRNA genes using Ribopicker.33 The remaining reads were then used for the BLASTX against the NR database at the “FutureGrid Portal (https://portal.futuregrid.org/)”. The output text file of the BLASTX was imported into MEGAN,34 to carry out phylogenetic, and KEEG analysis. The MEGAN phylogenetic output file was used to present the relative abundance of transcripts belonging to different bacterial and archaeal groups (Fig. 1).Mapping mRNA reads
For analysis of gene expression in Methanosaeta, digester mRNA reads were mapped against the published genome of Methanosaeta concilii (NC_015416.1), the dominant Methanosaeta in the digesters.19Geobacter gene expression was examined by mapping against the genome of G. metallireducens (NC_007517.1). Mapped reads were normalized with the RPKM (reads assigned per kilobase of target per million mapped reads) method35 using ARRAY STAR. Co-culture sequence reads were filtered for mRNA sequences, which were then mapped against G. metallireducens (NC_007517.1) and M. harundinacea (NC_017527) genomes as described previously.12 Reads from biological replicates were compared with each other graphically after mapping onto the template genomes (Table S1, Fig. S2†). Due to high reproducibility of data from biological replicates (Fig. S2†) the values were merged and averaged before further analysis (Tables S2 and S3†).Accession number
Sequence reads have been submitted to the EMBL databases under accession no. ERP003805.Conclusions
The results demonstrate that Methanosaeta species can directly accept electrons through biological electrical connections for the reduction of carbon dioxide to methane and that DIET can predominate over interspecies H2/formate transfer during anaerobic digestion. These findings greatly expand the known metabolic capabilities of Methanosaeta, which are abundant not only in anaerobic digesters,19,20 but also in a diversity of methanogenic soils and sediments.36–39Methanosaeta is considered to produce more methane on Earth than any other methanogen, due to its ubiquitous distribution and its high affinity for acetate, the precursor of more than half of the methane in most methanogenic environments.21 However, the energy yield from the conversion of acetate to methane is low (−75.7 kJ mol−1 methane) and the ability of Methanosaeta to also produce methane with electrons derived from DIET may add to their competitive advantage. The electron-accepting components that allow Methanosaeta species to participate in DIET are not known. However, the apparent ability of other methanogens to accept electrons from abiotic donors such as metallic iron40,41 or electrodes,42 as well as the enhancement of electron transfer between Geobacter and Methanosarcina species with conductive minerals43 or granular activated carbon44 is analogous to the biological connections proposed here. The inability of Methanospirillum hungatei or Methanobacterium formicicum to form co-cultures with G. metallireducens suggests that not all methanogens are capable of DIET.The results demonstrate that although Geobacter species are primarily known for their ability to grow with the reduction of extracellular electron acceptors, such as Fe(III) oxides, humic substances, and electrodes,45 they are also effective syntrophs, essentially using other organisms as another extracellular electron sink. Geobacter species were the most metabolically active microorganisms in methanogenic rice paddy soils,46 which suggests they may function as syntrophs in methanogenic environments other than anaerobic digesters. There is a wide diversity of organisms that are known to grow in co-culture with methanogens via interspecies H2/formate transfer,4,8–10 which when grown with Methanosaeta, may also be found to be capable of DIET.
The importance of DIET in a diversity of methanogenic environments is as yet unknown. Analysis of twenty-four brewery waste digesters revealed that each produced electrically conductive aggregates in which Geobacter and Methanosaeta species were abundant, suggesting that DIET is common in such systems.20 It should be possible to determine the relative importance of DIET in digesters treating more complex wastes as well as in methanogenic soils and sediments with the metatranscriptomic approach described here.
Acknowledgements
Authors wish to acknowledge laboratory assistance from Trevor Woodard, Joy Ward, and Beatrice Markovaite. This research was supported by the Office of Science (BER), U.S. Department of Energy, award no. DE-SC0004485.Notes and references
- P. L. McCarty, J. Bae and J. Kim, Environ. Sci. Technol., 2011, 45, 7100–7106 CrossRef CAS PubMed.
- M. Madsen, J. B. Holm-Nielsen and K. H. Esbensen, Renewable Sustainable Energy Rev., 2011, 15, 3141–3155 CrossRef CAS PubMed.
- M. P. Bryant, E. A. Wolin, M. J. Wolin and R. S. Wolfe, Arch. Mikrobiol., 1967, 59, 20–31 CAS.
- J. R. Sieber, M. J. McInerney and R. P. Gunsalus, Annu. Rev. Microbiol., 2012, 66, 429–452 CrossRef CAS PubMed.
- A. J. Stams and C. M. Plugge, Nat. Rev. Microbiol., 2009, 7, 568–577 CrossRef CAS PubMed.
- J. Dolfing, B. Jiang, A. M. Henstra, A. J. Stams and C. M. Plugge, Appl. Environ. Microbiol., 2008, 74, 6126–6131 CrossRef CAS PubMed.
- F. A. de Bok, M. L. Luijten and A. J. Stams, Appl. Environ. Microbiol., 2002, 68, 4247–4252 CrossRef CAS.
- B. E. Jackson, V. K. Bhupathiraju, R. S. Tanner, C. R. Woese and M. J. McInerney, Arch. Microbiol., 1999, 171, 107–114 CrossRef CAS.
- M. J. McInerney and M. P. Bryant, Appl. Environ. Microbiol., 1981, 41, 346–354 CAS.
- R. Cord-Ruwisch, D. R. Lovley and B. Schink, Appl. Environ. Microbiol., 1998, 64, 2232–2236 CAS.
- Z. M. Summers, H. E. Fogarty, C. Leang, A. E. Franks, N. S. Malvankar and D. R. Lovley, Science, 2010, 330, 1413–1415 CrossRef CAS PubMed.
- P. M. Shrestha, A.-E. Rotaru, Z. M. Summers, M. Shrestha, F. Liu and D. R. Lovley, Appl. Environ. Microbiol., 2013, 79, 2397–2404 CrossRef CAS PubMed.
- P. M. Shrestha, A. E. Rotaru, M. Aklujkar, F. Liu, M. Shrestha, Z. M. Summers, N. Malvankar, D. C. Flores and D. R. Lovley, Environ. Microbiol. Rep., 2013, 1–7 Search PubMed.
- D. Lovley, Rev. Environ. Sci. Biotechnol., 2011, 10, 101–105 CrossRef.
- A. E. Rotaru, P. M. Shrestha, F. Liu, T. Ueki, K. Nevin, Z. M. Summers and D. R. Lovley, Appl. Environ. Microbiol., 2012, 78, 7645–7651 CrossRef CAS PubMed.
- G. Reguera, K. D. McCarthy, T. Mehta, J. S. Nicoll, M. T. Tuominen and D. R. Lovley, Nature, 2005, 435, 1098–1101 CrossRef CAS PubMed.
- N. S. Malvankar, M. Vargas, K. P. Nevin, A. E. Franks, C. Leang, B. C. Kim, K. Inoue, T. Mester, S. F. Covalla, J. P. Johnson, V. M. Rotello, M. T. Tuominen and D. R. Lovley, Nat. Nanotechnol., 2011, 6, 573–579 CrossRef PubMed.
- M. Vargas, N. S. Malvankar, P.-L. Tremblay, C. Leang, J. A. Smith, P. Patel, O. Synoeyenbos-West, K. P. Nevin and D. R. Lovley, mBio, 2013, 4, e00105 Search PubMed.
- M. Morita, N. S. Malvankar, A. E. Franks, Z. M. Summers, L. Giloteaux, A. E. Rotaru, C. Rotaru and D. R. Lovley, mBio, 2011, 2, e00159 CrossRef CAS PubMed.
- P. Shrestha, N. Malvankhar, A. E. Rotaru, A. E. Franks, M. Shrestha, D. Shrestha, F. Liu, K. P. Nevin and D. R. Lovley, submitted.
- K. S. Smith and C. Ingram-Smith, Trends Microbiol., 2007, 15, 150–155 CrossRef CAS PubMed.
- K. Ma, X. Liu and X. Dong, Int. J. Syst. Evol. Microbiol., 2006, 56, 127–131 CrossRef CAS PubMed.
- H. Nagarajan, M. Embree, A. E. Rotaru, P. M. Shrestha, A. M. Feist, B. O. Palsson, D. R. Lovley and K. Zengler, Nat. Commun. Search PubMed , in Press.
- J. Zhu, H. Zheng, G. Ai, G. Zhang, D. Liu, X. Liu and X. Dong, PLoS One, 2012, 7, e36756 CAS.
- P.-L. Tremblay, M. Aklujkar, C. Leang, K. P. Nevin and D. Lovley, Environ. Microbiol. Rep., 2011, 4, 82–88 CrossRef PubMed.
- M. V. Coppi, C. Leang, S. J. Sandler and D. R. Lovley, Appl. Environ. Microbiol., 2001, 67, 3180–3187 CrossRef CAS PubMed.
- J. D. Coates, J. Woodward, J. Allen, P. Philp and D. R. Lovley, Appl. Environ. Microbiol., 1997, 63, 3589–3593 CAS.
- J. Pernthaler, F. O. Glockner, W. Schonhuber and R. Amann, in Methods in microbiology: marine microbiology, ed. J. Paul, Academic Press Ltd., London, 2001 Search PubMed.
- L. Raskin, D. Zheng, M. E. Griffin, P. G. Stroot, P. Misra and A. Van Leeuw, J. Microbiol., 1995, 68, 297–308 CAS.
- K. L. Lowe, T. J. Dichristina, A. N. Roychoudhury and P. Van Cappellen, Geomicrobiol. J., 2000, 17, 163–178 CrossRef CAS.
- J. Z. Levin, M. Yassour, X. Adiconis, C. Nusbaum, D. A. Thompson, N. Friedman, A. Gnirke and A. Regev, Nat. Methods, 2010, 7, 709–715 CrossRef CAS PubMed.
- R. Schmieder and R. Edwards, Bioinformatics, 2011, 27, 863–864 CrossRef CAS PubMed.
- R. Schmieder, Y. W. Lim and R. Edwards, Bioinformatics, 2012, 28, 433–435 CrossRef CAS PubMed.
- D. H. Huson, A. F. Auch, J. Qi and S. C. Schuster, Genome Res., 2007, 17, 377–386 CrossRef CAS PubMed.
- A. Mortazavi, B. A. Williams, K. McCue, L. Schaeffer and B. Wold, Nat. Methods, 2008, 5, 621–628 CrossRef CAS PubMed.
- K. Z. Falz, C. Holliger, R. Grosskopf, W. Liesack, A. Nozhevnikova, B. Muller, B. Wehrli and D. Hahn, Appl. Environ. Microbiol., 1999, 65, 2402–2408 CAS.
- A. Fey and R. Conrad, Appl. Environ. Microbiol., 2000, 66, 4790–4797 CrossRef CAS.
- S. Beckmann, T. Lueders, M. Kruger, F. von Netzer, B. Engelen and H. Cypionka, Appl. Environ. Microbiol., 2011, 77, 3749–3756 CrossRef CAS PubMed.
- Y.-Q. Tang, Y. Li, J.-Y. Zhao, C.-Q. Chi, L.-X. Huang, H.-P. Dong and X.-L. Wu, PLoS One, 2012, 7, e33535 CAS.
- H. T. Dinh, J. Kuever, M. Mussmann, A. W. Hassel, M. Stratmann and F. Widdel, Nature, 2004, 427, 829–832 CrossRef CAS PubMed.
- T. Uchiyama, K. Ito, K. Mori, H. Tsurumaru and S. Harayama, Appl. Environ. Microbiol., 2010, 76, 1783–1788 CrossRef CAS PubMed.
- S. Cheng, D. Xing, D. F. Call and B. E. Logan, Environ. Sci. Technol., 2009, 43, 3953–3958 CrossRef CAS.
- S. Kato, K. Hashimoto and K. Watanabe, Environ. Microbiol., 2012, 14, 1646–1654 CrossRef CAS PubMed.
- F. Liu, A. E. Rotaru, P. M. Shrestha, N. S. Malvankar, K. P. Nevin and D. R. Lovley, Energy Environ. Sci., 2012, 5, 8982–8989 CAS.
- D. R. Lovley, T. Ueki, T. Zhang, N. S. Malvankar, P. M. Shrestha, K. A. Flanagan, M. Aklujkar, J. E. Butler, L. Giloteaux, A. E. Rotaru, D. E. Holmes, A. E. Franks, R. Orellana, C. Risso and K. P. Nevin, Adv. Microb. Physiol., 2011, 59, 1–100 CrossRef CAS PubMed.
- T. Hori, M. Noll, Y. Igarashi, M. W. Friedrich and R. Conrad, Appl. Environ. Microbiol., 2007, 73, 101–109 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) is available. See DOI: 10.1039/c3ee42189a |
| ‡ Authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2014 |