Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Atomic force microscopy in biomaterials surface science
Fabio
Variola
*ab
aFaculty of Engineering, Department of Mechanical Engineering, University of Ottawa, Ottawa, ON K1N 6N5, Canada. E-mail: Fabio.variola@uottawa.ca
bFaculty of Science, Department of Physics, University of Ottawa, Ottawa, ON K1N 6N5, Canada
First published on 10th December 2014
Abstract
Recent progress in surface science, nanotechnology and biophysics has cast new light on the correlation between the physicochemical properties of biomaterials and the resulting biological response. One experimental tool that promises to generate an increasingly more sophisticated knowledge of how proteins, cells and bacteria interact with nanostructured surfaces is the atomic force microscope (AFM). This unique instrument permits to close in on interfacial events at the scale at which they occur, the nanoscale. This perspective covers recent developments in the exploitation of the AFM, and suggests insights on future opportunities that can arise from the exploitation of this powerful technique.
Dr Fabio Variola received his MSc in Materials Engineering from the University of Trieste. He then completed a joint doctoral program between the Institut National de la Recherche Scientifique-Énergie, Matériaux et Télécommunications (INRS-ÉMT) and the Université de Montréal. Since 2010, Dr F. Variola has worked as an Assistant Professor in the Department of Mechanical Engineering/Physics at the University of Ottawa. His expertise conjugates materials engineering with surface science and nanotechnology. As an independent investigator, Dr F. Variola has focused on understanding the cellular and molecular mechanisms that dictate how mammalian cells and bacteria respond to biomaterials. |
1. Introduction
Important scientific discoveries have always followed closely the technological development of novel tools for observing natural and physical phenomena at various scales. For example, the evolution of the telescope, from Galileo's first astronomical observations to the latest evidence supporting the Big Bang theory, has propelled our knowledge of celestial events. At the other end of the scale, progress in microscopy techniques has revealed increasingly smaller features of matter, ranging from the finest structures of plants and biological tissues to single atoms.1 Among the various instruments that enabled to resolve what is invisible to the eye, the atomic force microscope (AFM) has rapidly emerged as one of the most efficient and flexible tools to probe materials at the micro- and nanoscales.2 One of the disciplines that has benefitted the most from this technique is undoubtedly biomaterials surface science, a research field that aims at elucidating the interactions between materials and the surrounding biological environment by synergistically integrating different branches of science such as nanotechnology, biology, chemistry and physics. In this context, the AFM has contributed significantly to determining that the activity of proteins, cells and bacteria is affected by the nanometric physicochemical features of surfaces.3–8 While the main focus of this perspective will be the nanoscale interactions between biological entities and synthetic biomaterials (i.e. abiotic materials intended to interact with biological systems), it should be noted that the AFM has also been advantageously exploited to probe the effects of the microscale surface properties on biological activity7,9,10 and to investigate naturally occurring biomaterials (e.g. bone, teeth, cartilages, collagen and biomembranes).11–15Concisely, the AFM allows to image, measure (quantify), manipulate and sense matter at the nanometric level. From this first description, it readily transpires that this microscope is not simply an instrument to visualize small features, but instead a more powerful toolbox that offers additional remarkable capacities beyond imaging. These will be discussed below, aiming at providing the reader with a critical overview of selected applications of the AFM in biomaterials surface science.
2. AFM basic principles
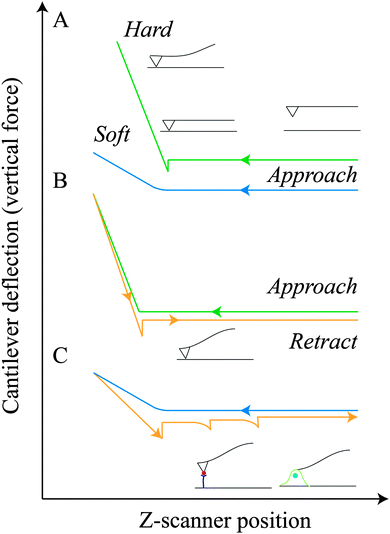
The AFM consists of a sharp tip mounted at the end of a microfabricated flexible cantilever which is raster scanned across the sample's surface. When the tip is brought in close proximity to the sample surface, attractive and repulsive forces cause the deflection of the cantilever. This deflection is detected and processed as a function of the position on the (x, y) plane in order to obtain topographical images. The AFM can operate in a variety of modes, including contact (the tip is in continuous contact with the surface), non-contact (the cantilever vibrates and variations from its resonance frequency are used to generate images) and intermittent (the cantilever moves rapidly with a large oscillation between the repulsive and attractive forces).16 If, in place of scanning, the tip is placed at a fixed point and moved vertically towards the sample and then retracted (force–distance analysis), the deflection of the cantilever will provide information on the mechanical properties of the surface, such as stiffness (as the tip approaches and indents the sample) and adhesion forces (as the tip tries to disengage from the sample) (Section 3.4).16 For hard materials (e.g. metals, ceramics) the cantilever will simply approach the surface, jump into contact and then deflect (Fig. 1A, green line). For softer samples (e.g. polymers, cells) the curve will resemble the blue one in Fig. 1A. If adhesion forces generate between the tip and the surface, the cantilever will deflect downwards as it retracts away from the sample, jumping back to its initial position as the maximum pulling force of the cantilever exceeds the tip-sample adhesion (Fig. 1B, orange line).17 If a molecule or a single cell is coupled with the tip (Section 3.5), as the cantilever moves away from the sample, molecule–surface, molecule–molecule (e.g. ligand–receptor), cell–cell and cell–surface adhesion bonds will be progressively ruptured throughout a series of unbinding events until full detachment, generating the characteristics “jumps” in the force–displacement curve (Fig. 1C, orange curve). For a more exhaustive coverage of the AFM basics, readers are invited to consult the comprehensive literature on the subject.16,173. Major applications of atomic force microscopy
3.1 Imaging
It is now well-known that nanotopography is one of the main parameters that governs protein adsorption onto biomaterials3 and dictates the interfacial interactions with cells and bacteria.4,8 Therefore, a precise 3-dimensional characterization of the nanometric features of surfaces is a fundamental step for understanding the biological response to nanostructured biomaterials. AFM conjugates high spatial resolution with experimental flexibility (i.e. it can be applied to diverse conductive and non-conductive materials, and requires little to no sample preparation), reasons that justify its widespread use not only to display the nanotopography of surfaces but also quantify their micro and nanoscale roughness.18–24 Despite its flexibility and advantages, this technique can be susceptible to artifacts that may lead to the erroneous interpretation of data. For instance, the actual profile of sharp vertical steps can be altered by the tip geometry and/or the radius of the tip itself.16 Similarly, the height/depth of porous/hollow structures (e.g. 3-dimensional scaffolds for tissue engineering and nanoporous platforms for drug-delivery applications), for which the full penetration of the tip is hampered by its geometry, may be underestimated. While these limitations can be overcome by using sharp and ultra-sharp tips,16,25 there is still a trade-off between the image quality and the mechanical strength of the tip itself. Sharp tips are in fact more prone to blunt and/or break, which can lead to additional artifacts.16The AFM is not only capable of precisely resolving the nanotopography of surfaces, but also of mapping the spatial distribution of their physicochemical properties, such as, for instance, charge density and potential (i.e. Kelvin-probe force microscopy, a technique that detects and measures the electrostatic interactions between a conductive AFM tip and the substrate).26,27 In this context, the electrical properties of surfaces are critical factors in affecting cell–substrate interactions.28,29 It was in fact shown that nanotopographical surfaces influence charge density and electric field strength, and, in turn, the protein-mediated adhesion of cells.28,30 In particular, it was proposed that positively charged proteins (or proteins with positively charged domains) are responsible for dictating the subsequent attachment of negatively charged cells onto a negatively charged nanorough surface.29 While the majority of these studies employed computational methods to elucidate proteins and cell behavior on such surfaces, only a few exploited experimental approaches to validate the proposed analytical models and mechanisms.31 To this end, the capacity of AFM to visualize nanotopographical features and correlate them to surface charge density and potential26,27 will offer interesting new opportunities to consolidate the present knowledge of the relationships between the nanotopography and electrical characteristics of biomaterials, and ultimately determine how these influence proteins, cells and bacteria functions.
3.2 AFM and other imaging techniques
In general, compared to other techniques such as standard Scanning (SEM) and Transmission Electron (TEM) microscopy, the AFM not only offers the capacity to visualize nanometric features in two and three dimensions (for significantly smaller areas though, when compared to SEM) and to map the physicochemical characteristics of surfaces, but it also advantageously permits to do so in air, vacuum and especially in liquid environments. These characteristics have been fundamental to image static and dynamic events, such as the adsorption of proteins and DNA,32–36 as well as the structural and molecular components of living cells and bacteria (Fig. 2),37–44 while providing minimal perturbation of the native characteristics of biological samples and avoiding artifacts associated with drying, including the creation of artefactual structures (e.g. dendritic patterns resulting from the crystallization of the buffer).16 In this context, for cells and bacteria imaging, chemical immobilization is often required to anchor them onto substrates in order to prevent their movement during imaging (common methods include fixation with glutaraldehyde and adsorption onto surfaces pre-coated with polymers, among others).39,45 This procedure may however alter the nature of interfacial interactions,46 thereby posing a challenge for studies aimed at assessing the direct effects of specific surface cues on the biological response. Standard fixation procedures that involve proteins crosslinking can also perturb the natural properties of cell wall architecture,46 and, in turn, may affect nanomechanical measurements (Section 3.4). The attachment methods that result in the nearest to natural state for cells are those that do not involve any chemical treatment,45 such as physical entrapment in porous membranes, confinement in micro-wells and electrostatic interactions.39 Noteworthy, immobilization protocols for live imaging may not be always necessary if cells adhere onto nanostructured biomaterials. These have in fact shown to promote a firmer cellular adhesion in physiological conditions by direct physical cueing,47 and thus promise to provide the required adhesion strength for imaging without any external fixation protocol. In general, the AFM procedure for imaging living cells should be fast and only rely on very small interaction forces to prevent cells from moving, detaching and/or being damaged. In this context, intermittent AFM-based approaches permit to reduce the imaging time, minimizing the risk of movement of the cell during the scan.46,48 In particular, Digital Pulsed Force mode (DPFM) imparts sinusoidal movement to the cantilever and modulates its oscillation at high frequency to make consecutive tip indentations on the surface, ultimately yielding faster scanning speeds while exerting small and controlled interaction forces.48,49 These and similar results (Section 3.4) demonstrate that effective AFM-based approaches to image and assess the structural properties of living cells and bacteria are readily available, and undoubtedly offer new opportunities to investigate cellular functions and interactions with surfaces with minimal perturbation.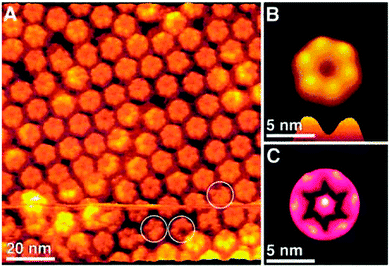 | ||
| Fig. 2 (A) AFM images of the arrangement of connexons (hexameric half-channels that bridge the extracellular space between adjacent plasma membranes) in gap junctions plaques. (B) Top and side view of a single connexon. (C) Enhanced structural details of a single connexon. Reprinted with permission from ref. 37. Copyright 2002 American Chemical Society. | ||
3.3 Chemical force microscopy
The already impressive imaging capacities have been even further expanded by integrating the AFM's spatial resolution and sensitivity with chemical functionalization, a set of strategies to immobilize molecular agents onto the cantilever's tip. The possibility of tailoring the AFM probe with chemical groups and individual molecules, coupled with the ability to carry out measurements under near-physiological conditions, made this approach ideal for biological applications.50 To date, chemical force microscopy (CFM) has been successfully exploited to visualize the distribution of molecules in surface-adsorbed protein layers51 and map the supramolecular organization of cellular membranes.52–55 These abilities also extend to the investigation of bacteria, and have permitted to visualize the structure and properties (e.g. hydrophobicity) of wall components (Fig. 3).39,56,57 Because of the abundance and flexibility of chemical strategies that can be tailored to precisely target selectively chosen features,58 tip functionalization and force imaging hold the potential not only to display, but also quantify (Section 3.5), a wide variety of relevant interfacial phenomena, such as the spatial distribution of specific binding sites on surfaces and its effects on the activity of adhering cells.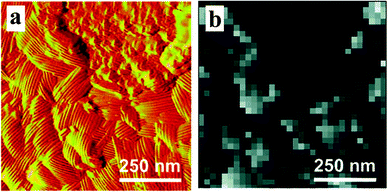 | ||
| Fig. 3 (a) AFM topographic image of the nanoscale structure of a microorganism (Aspergillus fumigatus) and (b) chemical force image of its hydrophobic domains. Reprinted with permission from ref. 56. Copyright 2007 American Chemical Society. | ||
3.4 Nanomechanical measurements
In parallel with imaging capacities, one of the distinctive features that has propelled the use AFM in nanosciences is undoubtedly its capability to quantify physicochemical properties of materials and biological entities. To date, AFM analysis has yielded a deeper understanding of the nanoscale mechanical properties of materials, proteins, cells and bacteria, supporting standard characterization and microscopy techniques (e.g. optical and fluorescence microscopy) with quantitative data. Advances that stemmed from such approach will be presented in more detail below, providing further evidence of the power of the AFM as a multifunctional nanoscale tool.A growing number of studies has been exploring the role of micro- and nano-level mechanics on the cellular response to surfaces.59–62 For example, it is now well known that adhering cells sense and respond to the substrate's stiffness.63 In this context, the AFM's capacity to quantify the stiffness and the hardness of natural and synthetic materials has provided important new resources for materials scientists interested in the connections between the nanomechanical characteristics of surfaces and the physicochemical properties of bulk materials.49,64 In fact, compared to standard techniques (e.g. tensile testing, micro-hardness, etc.), the AFM distinctively offers a non-destructive way to probe multiple properties at once (i.e. stiffness, maximum indentation, indentation at maximum force, hysteresis, hardness, compliance, adhesion force, detachment distance, detachment energy, Young's modulus)49 with a nanometric spatial resolution, and to simultaneously overlap them to the morphological images obtained with one of the modes outlined in Section 2.
In parallel, biologists and biophysicists have capitalized on force–distance measurements (Section 2) to close in on the nanomechanical properties of cells and bacteria such as Young's modulus and deformation,46,65,66 and determined how these vary during adaptation to the extracellular environment and/or to external physicochemical stimuli.62,65,67–69 A particular attention has been given to assessing how mammalian cells translate mechanical cues into cytoskeletal changes and biochemical signaling, and how the mechanical properties of cells are involved in regulating cellular functions (mechanosensing, mechanotransduction, and mechanoresponse).69 In this context, intermittent techniques allowed to map the mechanical properties of cells and bacteria, offering the distinctive advantage of providing their spatial distribution within single cells. For example, the mapping and time evolution of the stiffness of cancer cells were successfully imaged by DPFM (Section 3.2).48 In parallel, a recent study exploited a similar quantitative AFM procedure that provides precise and continuous control on the acquisition velocity and vertical forces to investigate living bacteria.46 The bacterial suspension was gently deposited on a glass substrate and force distant curves were collected for every pixel of AFM images, simultaneously generating topography and stiffness maps without the need of any external immobilization step (Fig. 4).
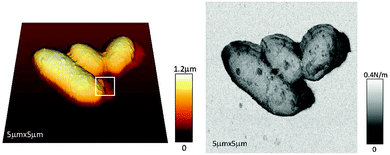 | ||
| Fig. 4 AFM height (left) and stiffness (right) images of bacteria (Rhodococcus wratislaviensis) in their physiological medium. Reprinted with permission from ref. 46. Copyright 2013, PLoS One. | ||
While these studies have undoubtedly provided a great contribution in advancing knowledge in their respective fields, they appear to have progressed along two sets of parallel lines, focused on either the nanomechanical properties of materials or on the biomechanics of biological entities. Ultimately, these two lines should converge towards the investigation of the mechanoresponse to the local physical/mechanical cueing exerted by nanostructures. In fact, it could be conceived that nanoscale surface features engender a precise nanomechanical environment capable of provoking localized cell-distortions, inducing biomechanical changes in cytoskeletal organization and cellular shape.7 Although further refinements are still needed, these phenomena can all be characterized by the AFM, which thus promises to support previous63 and ongoing efforts to bridge the gap between substrate and cellular mechanics. In this context, although the AFM has been successfully employed to quantify the stiffness of bacterial membrane,46 its application to study how bacteria respond to nanostructured surfaces70 from a nanomechanical viewpoint is still lagging behind. Once addressed, the generated knowledge is poised to bring new evidence to elucidate the mechanisms involved during the interactions between bacteria and nanostructured antibacterial surfaces.71–73
3.5 Force spectroscopy
In addition to force imaging, tip functionalization has also permitted to quantify, with unparalleled spatial (∼1 nm) and force (in the order of piconewtons) resolution, a remarkable collection of molecular, cellular and bacterial properties and functions (Force spectroscopy).53 In the force spectroscopy technique, the AFM tip is functionalized with chemical groups (i.e. chemical force spectroscopy-CFS), biomolecules, proteins and viruses (i.e. single-molecule force spectroscopy-SMFS), as well as with single living cells (i.e. single-cell force spectroscopy-SCFS), and interactive forces acting on the functionalized tip when scanned across a sample are measured.39 This approach has provided direct access to a variety of inter- and intramolecular phenomena and cellular events otherwise inaccessible with other techniques. For example, single-molecule force spectroscopy quantified via force–distance curves protein unraveling, folding and unfolding mechanisms, receptor–ligand and ligand-binding interactions.53,74,75 Research aiming at characterizing protein–biomaterials interactions can capitalize on these and similar studies76 and determine how nanometric surface cues affect parameters of protein adsorption such as adhesion and unbinding forces. The capacity of SMFS to probe molecular events can also be exploited to investigate the interactions and the dynamics of a variety of cell–surface-associated proteins.77 This also extends to bacteria research, where this technique has enabled a better understanding of the molecular mechanisms underlying the adhesive and mechanical properties of polysaccharides and proteins on live cells.78,79 Based on these studies, researchers are offered the opportunity to exploit SMFS to dissect out specific aspects of the molecular bases of cellular and bacteria adhesion processes on nanostructured biomaterials, to determine, for instance, whether the physicochemical cueing exerted by surface cues also affects the supramolecular architecture of adhering cells.A field that will particularly benefit from advancements in SMFS is drug-delivery, particularly its branch that employs nanostructured biomaterials for localized and controlled elution of antibiotics, growth factors and viruses.80–82 In fact, the type and strength of the specific interactions established by bioactive molecules with surfaces are expected to impact their tendency to desorb, ultimately dictating the overall release profile.18 While the ability of the AFM to quantify the strength of adhesion forces can nonetheless provide important guidance for assessing the quality of molecule–surface interactions, care should be taken to relate the measured forces to the binding affinity, which was shown to depend to a large extend on the experimental parameters (e.g. contact time).76 In this context, various groups have been exploring the controlled delivery of viral vectors to overcome some of the current challenges in gene therapy and tissue engineering by improving site targeting, reducing inflammatory and immune responses, enhancing transduction and extending the duration of gene expression.82 Recently, strategies for specific vector-biomaterial binding based on the interactions of functional groups on viral capsid and functionalized surfaces (e.g. with antiviral antibodies) have been developed.82 The main goal is devising flexible approaches that permit to tailor the release and delivery to meet specific therapeutic needs by tuning the vector affinity for the surface. To this end, researchers can capitalize on previous work which exploited the AFM to determine how the structural and nanomechanical properties of viruses (e.g. stiffness of the viral shell),82–87 as well as their binding strength with materials, quantified by SMFS for instance, are affected by the physiochemical environment of surfaces. The AFM thus promises to become an essential tool also in the field of substrate-mediated viral gene delivery that will permit to make the correlation between the nature and quality of the initial virus–surface interactions and the resulting elution profiles. This will provide the key for optimizing vector-presenting substrates and design better performing drug-eluting platforms capable of retaining and progressively releasing the virus for cellular internalization. In addition, tuning surface properties to control vector–material interactions is also expected to unlock new strategies to preserve vector activity and avoid recognition by the immune system.82
At the cellular level, single cells can be ingeniously fixed onto the cantilever, becoming the probe for the dynamic quantification of cell–substrate interactions (SCFS).88,89 Parameters such as maximum detachment force and work for the entire cells as well as a number of detachment events of single cellular tethers have already been successfully quantified (Fig. 5).88,89 It should be noted that this technique still faces experimental challenges and limitations.88 For example, immobilization strategies have to ensure a firm attachment of individual cells onto the cantilever in ways that the cantilever-cell adhesion strength will be greater than the cell–substrate interactions to prevent the cell from adhering onto the substrate during the retraction of the tip. It follows that nanostructured materials that promote a strong cellular adhesion may cause the detachment of the cell from the cantilever. In addition, SCFS experiments are time consuming because only one cell can be characterized at a time. For statistical reasons, many detachment-force–distance curves must be recorded, which limits the length of the contact times that can reasonably be assayed, often restricting the analysis to short contact times (from milliseconds to about 20 minutes).88 Despite these challenges, SCFS nonetheless provides unique insight into the forces, energetics and kinetics of cell adhesion processes onto surfaces.88 In this context, it will be interesting to move the focus from whole cells to more specialized cellular components, and complement existing techniques (e.g. optical tweezers)90 with AFM-based approaches to investigate the local properties of filopodia emitted by cells adhering on nanostructured surfaces. A better understanding of the relationship of filopodia with surface features at their functional sensing scale, that is the nanoscale, will be in fact highly relevant for exploiting nanotopographical surfaces to stimulate (i.e. regeneration) and even slowdown (i.e. cellular hyperplasia, cancer cells) cell activity.
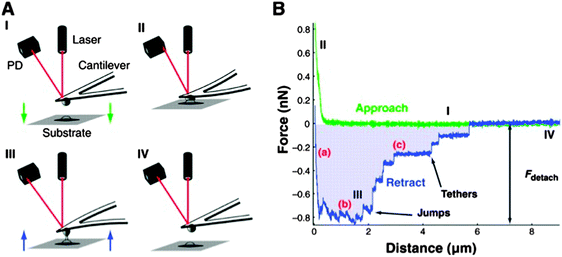 | ||
| Fig. 5 (A) Schematic overview of AFM-based single cell adhesion experiments. The cantilever-cell system approaches the surface (I) until contact is established (II). During detachment (III–IV), force distance curves are recorded. (B) Force–distance curve showing maximum detachment force (Fdetach), work of cell detachment from the substrate (shaded area) and two types of molecular unbinding events: (i) the receptor remains anchored in the cell cortex and unbinds as the force increases (denoted as jumps) and (ii) receptor anchoring is lost and membrane tethers are pulled out of the cell. Reprinted with permission from ref. 88. Copyright 2008, Company of Biologists. | ||
While single cell force spectroscopy has been implemented for studies involving mammalian cells, its application to bacterial research has not evolved as rapidly, mainly as a consequence of difficulties in immobilizing bacteria onto cantilevers. However, a recent article has reported the use of a cell adhesive to attach a single bacterium onto the cantilever and probe its interactions with different surfaces and other bacteria,91 thereby opening the window on additional possibilities to exploit the AFM to comprehend the mechanisms involved in the bacterial attachment to nanostructured surfaces.
To conclude, AFM force imaging and spectroscopy have permitted to achieve, through quantitative measurements of an unparalleled resolution, a deeper understanding of numerous molecular processes that underlay proteins, cells and bacteria activities. These techniques are not solely limited to biomaterials surface science, but their potential extend to other fields of science, ranging from drug delivery to medicine, where the capacity to quantify structures and interactions at the molecular level can contribute, for instance, to devise better performing pharmacologic strategies and elucidate the mechanisms of specific diseases.
3.6 Scanning probe lithography
In a further extension of AFM-based methods and techniques, the distinctive capacity to manipulate single molecules and detect their interactions has permitted researchers to explore additional applications in nanosciences, ultimately developing protocols and practices that harnessed the power of the AFM probe and transformed it into a multifunctional molecular toolbox.53 One of the most relevant techniques that stemmed from such a development falls under the umbrella of scanning probe lithography, a set of nanofabrication strategies in which the tip is used to directly pattern substrates with a variety of molecular “inks”.16 Among these, dip-pen nanolithography has shown promising applications in biomaterials surface science.92 In this method, selected molecules are adsorbed by capillarity onto the AFM tip, which is successively used in contact-mode to transfer pre-determined molecular patterns to the surface with a resolution of 30–100 nm.16 For instance, microarrays composed by DNA-directed protein immobilization have been precisely patterned and utilized to investigate the recruitment of transmembrane receptors in living cells (Fig. 6).93 This technique has been optimized on atomically-flat and contaminant-free surfaces (e.g. gold and silicon), and it can now count on a dramatically improved patterning efficiency and flexibility in depositing different ink molecules.92 However, its translation to medically relevant materials and applications may still be far, mainly because of the non atomic planarity of most biomaterials and the practical need to pattern areas whose dimensions lay beyond the range of this technique. Nonetheless, the capacity of creating specific molecular patters will undoubtedly benefit more fundamental studies in biomaterials surface science, focused, for example, on furthering the investigation of cellular response to specific chemical cues (e.g. chemotaxis)94 and/or on the validation of functional molecular nanostructures.95,96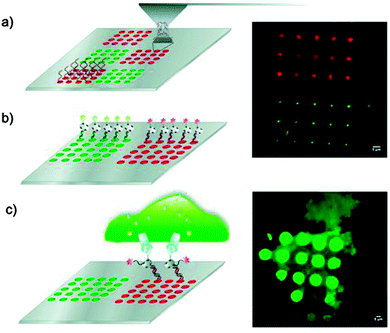 | ||
| Fig. 6 Schematic representation of the generation of microarrays for DNA-directed protein immobilization for the recruitment of transmembrane receptors (left, from top to bottom). Live-cell arrays (top right) and expression of transmembrane protein in breast cancer cells membrane (bottom right). Reprinted with permission from ref. 93. Copyright 2013, Wiley. | ||
3.7 Cantilever-based sensors
In parallel with these capacities, the AFM has also been employed to detect the deflection and/or resonance shift of cantilever arrays used as micro and nanomechanical sensor for diagnostic and detection purposes.97–100 In this context, researchers have demonstrated that silicon-based cantilevers can be functionalized with thin metallic (e.g. gold) and polymeric (e.g. polystyrene, PMMA) films to detect a variety of biological and chemical species.101–103 The knowledge acquired so far in the field may permit to change the identity of the deposited materials and explore functionalization strategies to create thin films of biocompatible materials with controlled properties directly on cantilever arrays, which could then be used to study protein adsorption, ultimately complementing techniques such as surface plasmon resonance (SPR, i.e. an optical technique sensitive to the changes in refractive index that occur when proteins and other molecules accumulate onto a gold surface) and quartz crystal microbalance (QCM, i.e. a physical technique that detects changes in the resonance frequency of an electrically driven quartz crystal with changes in mass due to molecule adsorption).104,1054. Outlook
The recent history of microscopy has witnessed a more systematic approach to the development and exploitation of novel strategies and protocols for the characterization of biomaterials and interfacial biological events at the micro- and nanoscales. In this context, the AFM has undoubtedly been a fundamental resource in biomaterials surface science to date, and will still play a major role in the future. In particular, the optimization of current tip functionalization approaches and the development of novel strategies for CFM, CFS, SMFS and SCFS will extend the breath of its applications and the specificity of the results, offering new opportunities to close in on still unknown aspects of how proteins, bacteria and cells sense and respond to nanostructured surfaces.However, there is still a number of experimental limitations that will need attention to take the technique to a higher level and further enhance its impact. For instance, structural AFM measurements on living cells/bacteria still pose experimental challenges, not only in regard to immobilization protocols (Section 3.2), but also in relation to sample damaging caused by forces exerted by the tip, imaging artifacts and interpretation of quantitative results. In addition, the time currently required to record a high-resolution image is much longer than the timescale of most biological processes.69 To address this need, the development and optimization of fast scanning AFMs and acquisition procedures (e.g. DPFM) for high-speed topographic imaging now permit to image the physical, chemical and biological properties of samples at high speed and resolution.68,69,106 Applied to biomaterials surface science, these new advances hold the potential to yield a dynamic description of the events at the molecular scale that affect and control cell–surface interactions.
In such a technological evolution towards more advanced and effective analyses, the AFM has joined a more general trend in microscopy which aims at multimodal imaging capacities and correlative analyses, becoming an instrument that can seamlessly integrate complementary characterization methods. This evolution has ultimately permitted to the AFM to achieve a synergy that extends beyond the capacities and potential of the constituent techniques used independently.
Standard and high-resolution fluorescence microscopy as well as Raman imaging and spectroscopy107–109 can all be combined within the AFM setup. Such advances now permit to achieving high-resolution images that simultaneously display the 3-dimensional topography of the cytoskeleton and its stiffness, and correlate them to its structural elements, such as the distribution of actin filaments visualized by standard confocal microscopy or by stimulated emission depletion microscopy (STED), a super high resolution technique that breaks the resolution limit of standard fluorescence microscopy (Fig. 7).110,111 This integrated approach is poised to unveil the connections between the cell nanomechanical properties and the organization of actin fibers and microtubules in the cellular body and its extensions, ultimately addressing, for instance, how the structural and mechanical parameters of cells and bacteria are affected by the physicochemical environment of surfaces. Similarly, when the AFM is coupled with a near-field scanning optical microscope (SNOM/NSOM), morphological and immunofluorescent information, such as the localization of molecular nanoclusters in cell membranes and protrusions (Fig. 8), can be overlapped.112–115 This promises to shed new light on the characteristics and variations of the cellular supramolecular architecture as a function of cell–substrate and cell–cell interactions on nanostructured biomaterials. The integration of Raman spectroscopy and imaging offers the distinctive advantage to carry out label-free chemical analyses of surfaces and adhering cells and bacteria,116,117 and successively correlate them to the nanotopography of the underlying nanostructures. Raman spectroscopy will also permit to probe, in a non-destructive manner, the crystallinity and even local stress fields of biomaterials,118–121 parameters known to affect cell and bacteria response.7,9,73 It should be noted that the spatial resolution of standard Raman imaging is dictated by the optical objective and the laser wavelength, and can achieve at best a few hundreds of nanometers, thus considerably above that of the AFM. Such a limitation can however be overcome by tip-enhanced Raman spectroscopy (TERS), a technique that significantly increases the resolution of Raman imaging and spectroscopy to a few nanometers.122 TERS have been used to investigate biomolecules, such as DNA bundles,123 and probe materials surfaces and biological samples at the nanoscale,124–128 thereby becoming a very promising technique for current and future studies in various disciplines.
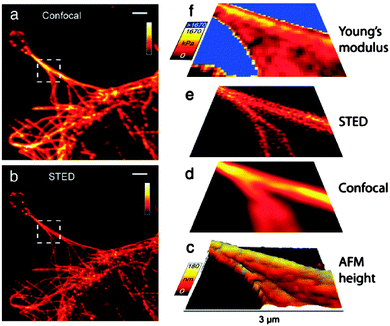 | ||
| Fig. 7 (a) Confocal image and (b) STED image [both raw data], (c) 3D rendered view extracted from AFM force curves; deconvolved (d) confocal image and (e) STED image, (f) elasticity map calculated from AFM force curves of fibroblastic cells. Scale bars in (a) and (b): 2 μm, axes bars in (c)–(f): 3 μm. Reprinted with permission from ref. 111. Copyright 2012, SpringerOpen. | ||
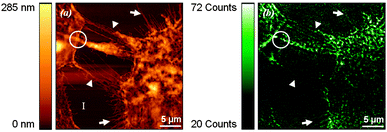 | ||
| Fig. 8 Topographic (left) and fluorescent (right) SNOM images of human epithelial cells initiating cell–cell contact. Protein (E-cadherin) clusters location is revealed. Reprinted with permission from ref. 113. Copyright 2012, Elsevier. | ||
The development of AFM-based integrated systems and, as a matter of fact, complementary multimodal instruments (e.g. Raman-SEM), is paving the way for a novel approach to applied research, in which the synergistic combination of different experimental techniques increasingly generates more efficient and flexible tools for the nanoscale characterization of synthetic and biological materials. The technical evolution that has propelled such correlative approach to science will also give new strength to biomaterials surface science, a field in which the AFM will continue to affirm its role as a unique resource.
Acknowledgements
F.V. thanks Drs Antonio Nanci and Shan Zou for critical reading of the manuscript and acknowledges funding from the Canada Foundation for Innovation (CFI), the Ontario Ministry of Research and Innovation, and the Natural Sciences and Engineering Research Council of Canada (NSERC).References
- C. Toumey, Nat. Nanotechnol., 2012, 7, 205–206 CrossRef CAS PubMed.
- P. Eaton and P. West, Atomic Force Microscopy, 2010 Search PubMed.
- M. S. Lord, M. Foss and F. Besenbacher, Nano Today, 2010, 5, 66–78 CrossRef CAS PubMed.
- M. J. Biggs, R. G. Richards and M. J. Dalby, Nanomedicine, 2010, 6, 619–633 CrossRef CAS PubMed.
- R. E. McMahon, L. Wang, R. Skoracki and A. B. Mathur, J. Biomed. Mater. Res., Part B, 2013, 101, 387–397 CrossRef PubMed.
- L.-C. Cheng, X. Jiang, J. Wang, C. Chen and R.-S. Liu, Nanoscale, 2013, 5, 3547–3569 RSC.
- F. Variola, J. B. Brunski, G. Orsini, P. T. d. Oliveira, R. Wazen and A. Nanci, Nanoscale, 2011, 3, 335–353 RSC.
- K. Anselme, P. Davidson, A. M. Popa, M. Giazzon, M. Liley and L. Ploux, Acta Biomater., 2010, 6, 3824–3846 CrossRef CAS PubMed.
- F. Variola, F. Vetrone, L. Richert, P. Jedrzejowski, J.-H. Yi, S. Zalzal, S. Clair, A. Sarkissian, D. F. Perepichka, J. D. Wuest, F. Rosei and A. Nanci, Small, 2009, 5, 996–1006 CrossRef CAS PubMed.
- J. Hasan, R. J. Crawford and E. P. Ivanova, Trends Biotechnol., 2013, 31, 295–304 CrossRef CAS PubMed.
- G. E. Fantner, O. Rabinovych, G. Schitter, P. Thurner, J. H. Kindt, M. M. Finch, J. C. Weaver, L. S. Golde, D. E. Morse, E. A. Lipman, I. W. Rangelow and P. K. Hansma, Compos. Sci. Technol., 2006, 66, 1205–1211 CrossRef CAS PubMed.
- G. E. Fantner, T. Hassenkam, J. H. Kindt, J. C. Weaver, H. Birkedal, L. Pechenik, J. A. Cutroni, G. A. Cidade, G. D. Stucky, D. E. Morse and P. K. Hansma, Nat. Mater., 2005, 4, 612–616 CrossRef CAS PubMed.
- Y. Lin and S. Xu, J. Microsc., 2011, 241, 291–302 CrossRef CAS PubMed.
- E. Rettler, S. Hoeppener, B. W. Sigusch and U. S. Schubert, J. Mater. Chem. B, 2013, 2789–2806 RSC.
- A. Alessandrini and P. Facci, Micron, 2012, 43, 1212–1223 CrossRef CAS PubMed.
- A. W. Coleman, A. N. Lazar, C. F. Rousseau, S. Cecillon and P. Shahgaldian, in Nanosystem characterization tools in the life sciences, ed. C. S. S. R. Kumar, Wiley-VCH, 2011 Search PubMed.
- G. Haugstad, Atomic Force Microscopy: understanding basic modes and advanced applications, Wiley, 2012 Search PubMed.
- A. Ketabchi, K. Komm, M. Miles-Rossouw, D. A. D. Cassani and F. Variola, PLoS One, 2014, 9, e92080–e92089 Search PubMed.
- J. Crear, K. M. Kummer and T. J. Webster, Int. J. Nanomed., 2013, 8, 995–1001 CrossRef PubMed.
- L. He, P. Zhao, Q. Han, X. Wang, X. Cai, Y. Shi, L. Zhou, Y. Zhang and W. Xue, Carbon, 2013, 56, 224–234 CrossRef CAS PubMed.
- C. Huang, M. Moosmann, J. Jin, T. Heiler, S. Walheim and T. Schimmel, Beilstein J. Nanotechnol., 2012, 3, 620–628 CrossRef CAS PubMed.
- P. A. George, K. Quinn and J. J. Cooper-White, Biomaterials, 2010, 31, 641–647 CrossRef CAS PubMed.
- Y. Wang, L. Zhang, L. Sun and T. J. Webster, Int. J. Nanomed., 2013, 8, 159–166 Search PubMed.
- N. J. Fredin, A. H. Broderick, M. E. Buck and D. M. Lynn, Biomacromolecules, 2009, 10, 994–1003 CrossRef CAS PubMed.
- J. Brown, P. Kocher, C. S. Ramanujan, D. N. Sharp, K. Torimitsu and J. F. Ryan, Ultramicroscopy, 2013, 133, 62–66 CrossRef CAS PubMed.
- K. Suzuki, K. Kobayashi, N. Oyabu, K. Matsushige and H. Yamada, J. Chem. Phys., 2014, 140, 054704 CrossRef PubMed.
- J. M. Pelto, S. P. Haimi, A. S. Siljander, S. S. Miettinen, K. M. Tappura, M. J. Higgins and G. G. Wallace, Langmuir, 2013, 29, 6099–6108 CrossRef CAS PubMed.
- E. Gongadze, D. Kabaso, S. Bauer, T. Slivnik, P. Schmuki, U. v. Rienen and A. Iglic, Int. J. Nanomed., 2011, 6, 1801–1816 Search PubMed.
- D. Kabaso, E. Gongadze, S. Perutkova, C. Matschegewski, V. Kralj-Iglic, U. Beck, U. v. Rienen and A. Iglic, Comput. Meth. Biomech. Biomed. Eng., 2011, 14, 469–482 CrossRef CAS PubMed.
- B. Kasemo, Surf. Sci., 2002, 500, 656–677 CrossRef CAS.
- I. O. Smith, M. J. Baumann and L. R. McCabe, J. Biomed. Mater. Res., Part A, 2004, 70, 436–441 CrossRef CAS PubMed.
- A. J. Katan and C. Dekker, Cell, 2011, 147, 979–982 CrossRef CAS PubMed.
- A. M. Whited and P. S.-H. Park, Biochim. Biophys. Acta, 2014, 1838, 56–68 CrossRef CAS PubMed.
- W. Kalle and P. Strappe, Micron, 2012, 43, 1224–1231 CrossRef CAS PubMed.
- A. Rajendran, M. Endo and H. Sugiyama, Adv. Protein Chem. Struct. Biol., 2012, 87, 5–55 CAS.
- A. Rajendran, M. Endo and H. Sugiyama, Chem. Rev., 2014, 114, 1493–1520 CrossRef CAS PubMed.
- D. J. Muller, G. Hand, A. Engel and G. Sosinsky, EMBO J., 2002, 21, 3598–3607 CrossRef CAS PubMed.
- D. J. Muller and Y. F. Dufrene, Trends Cell Biol., 2011, 21, 461–469 CrossRef PubMed.
- L. S. Dorobantu, G. G. Goss and R. E. Burrell, Micron, 2012, 43, 1312–1322 CrossRef CAS PubMed.
- F. Badique, D. R. Stamov, P. M. Davidson, M. Veuillet, G. n. Reiter, J.-N. Freund, C. M. Franz and K. Anselme, Biomaterials, 2013, 34, 2991–3001 CrossRef CAS PubMed.
- J. P. Bearinger, L. C. Dugan, L. Wu, H. Hill, A. T. Christian and J. A. Hubbell, BioTechniques, 2009, 46, 209–216 CrossRef CAS PubMed.
- A. Raman, S. Trigueros, A. Cartagena, A. P. Z. Stevenson, M. Susilo, E. Nauman and S. A. Contera, Nat. Nanotechnol., 2011, 6, 809–814 CrossRef CAS PubMed.
- A. Cartagena and A. Raman, Biophys. J., 2014, 106, 1033–1043 CrossRef CAS PubMed.
- H. Yamashita, A. Taoka, T. Uchihashi, T. Asano, T. Ando and Y. Fukumori, J. Mol. Biol., 2012, 422, 300–309 CrossRef CAS PubMed.
- H. K. Webb, V. K. Truong, J. Hasan, R. J. Crawford and E. P. Ivanova, J. Microbiol. Methods, 2011, 86, 131–139 CrossRef PubMed.
- S. Dhahri, M. Ramonda and C. Marlière, PLoS One, 2013, 8, e61663 CAS.
- P. Bertoncini, S. L. Chevalier, S. Lavenus, P. Layrolle and G. Louarn, J. Mol. Recognit., 2012, 25, 262–269 CrossRef CAS PubMed.
- O. Marti, M. Holzwarth and M. Beil, Nanotechnology, 2008, 19, 384015–384022 CrossRef PubMed.
- C. A. Rezende, L.-T. Lee and F. Galembeck, Langmuir, 2009, 25, 9938–9946 CrossRef CAS PubMed.
- K. C. Neuman and A. Nagy, Nat. Methods, 2008, 5, 491–505 CrossRef CAS PubMed.
- P. Soman, Z. Rice and C. A. Siedlecki, Micron, 2008, 39, 832–842 CrossRef CAS PubMed.
- J. J. Heinisch, P. N. Lipke, A. Beaussart, S. E. K. Chatel, V. Dupres, D. Alsteens and Y. F. Dufrêne, J. Cell Sci., 2012, 125, 4189–4195 CrossRef CAS PubMed.
- D. J. Muller and Y. F. Dufrene, Nat. Nanotechnol., 2008, 3, 261–269 CrossRef PubMed.
- V. Dupres, D. Alsteens, G. Andre and Y. F. Dufrêne, Trends Microbiol., 2010, 18, 397–405 CrossRef CAS PubMed.
- L. S. Dorobantu and M. R. Gray, Scanning, 2010, 32, 74–96 CrossRef CAS PubMed.
- E. Dague, D. Alsteens, J.-P. Latgé, C. Verbelen, D. Raze, A. R. Baulard and Y. F. Dufrêne, Nano Lett., 2007, 7, 3026–3030 CrossRef CAS PubMed.
- Y. F. Dufrene, J. Bacteriol., 2002, 184, 5205 CrossRef CAS.
- C. D. Blanchette, A. Loui and T. V. Ratto, in Handbook of Molecular Force Spectroscopy, ed. A. Noy, Springer, 2005 Search PubMed.
- G. Chen, Y. Lv, P. Guo, C. Lin, X. Zhang, L. Yang and Z. Xu, Curr. Stem Cell Res. Ther., 2013, 8, 313–323 CrossRef CAS.
- C.-M. Cheng, R. L. S. Jr. and P. R. LeDuc, J. Biomech., 2009, 42, 187–192 CrossRef PubMed.
- A. J. Engler, P. O. Humbert, B. Wehrle-Haller and V. M. Weaver, Science, 2009, 324, 208–212 CrossRef CAS PubMed.
- D. E. Discher, D. J. Mooney and P. W. Zandstra, Science, 2009, 324, 1673–1677 CrossRef CAS PubMed.
- D. E. Discher, P. Janmey and Y.-l. Wang, Science, 2005, 310, 1139–1143 CrossRef CAS PubMed.
- J. Landoulsi and V. Dupres, Phys. Chem. Chem. Phys., 2013, 15, 8429–8440 RSC.
- C. Heu, A. Berquand, C. Elie-Caille and L. Nicod, J. Struct. Biol., 2012, 178, 1–7 CrossRef CAS PubMed.
- D. Kirmizis and S. Logothetidis, Int. J. Nanomed., 2010, 5, 137–145 CrossRef.
- J. Li, D. Han and Y.-P. Zhao, Sci. Rep., 2014, 4, 3910–3920 Search PubMed.
- I. Medalsy, U. Hensen and D. J. Muller, Angew. Chem., Int. Ed., 2011, 50, 12103–12108 CrossRef CAS PubMed.
- Y. F. Dufrene and A. E. Pelling, Nanoscale, 2013, 5, 4094–4104 RSC.
- L. Rizzello, R. Cingolani and P. P. Pompa, Nanomedicine, 2013, 8, 807–821 CrossRef CAS PubMed.
- S. D. Puckett, E. Taylor, T. Raimondo and T. J. Webster, Biomaterials, 2010, 31, 706–713 CrossRef CAS PubMed.
- B. Ercan, E. Taylor, E. Alpaslan and T. J. Webster, Nanotechnology, 2011, 22, 295102 CrossRef PubMed.
- F. Variola, S. F. Zalzal, A. Leduc, J. Barbeau and A. Nanci, Int. J. Nanomed., 2014, 9, 2319–2325 CrossRef PubMed.
- P. J. Bujalowski and A. F. Oberhauser, Methods, 2013, 60, 151–160 CrossRef CAS PubMed.
- C.-K. Lee, Y.-M. Wang, L.-S. Huang and S. Lin, Micron, 2007, 38, 446–461 CrossRef CAS PubMed.
- L. Richert, A. Boukari, S. Berner, M. Dard and J. Hemmerlé, J. Biomater. Nanobiotechnol., 2011, 2, 244–249 CrossRef CAS.
- D. J. Müller, M. Krieg, D. Alsteens and Y. F. Dufrêne, Curr. Opin. Biotechnol., 2009, 20, 4–13 CrossRef PubMed.
- G. Francius, D. Alsteens, V. Dupres, S. Lebeer, S. D. Keersmaecker, J. V.-. den and Y. F. Dufrene, Nat. Protoc., 2009, 4, 939–946 CrossRef CAS PubMed.
- C. Berne, X. Ma, N. A. Licata, B. R. A. Neves, S. Setayeshgar, Y. V. Brun and B. Dragnea, J. Phys. Chem. B, 2013, 117, 10492–10503 CrossRef CAS PubMed.
- A. C. A. Wan and J. Y. Ying, Adv. Drug Delivery Rev., 2010, 62, 731–740 CrossRef CAS PubMed.
- L. Yang and T. J. Webster, Expert Opin. Drug Delivery, 2009, 6, 851–864 CrossRef CAS PubMed.
- J.-H. Jang, D. V. Schaffer and L. D. Shea, Mol. Ther., 2011, 19, 1407–1415 CrossRef CAS PubMed.
- M. Hernando-Perez, E. Pascual, M. Aznar, A. Ionel, J. R. Caston, A. Luque, J. L. Carrascosa, D. Reguera and P. J. d. Pablo, Nanoscale, 2014, 6, 2702–2709 RSC.
- P. Ni, Z. Wang, X. Ma, N. C. Das, P. Sokol, W. Chiu, B. Dragnea, M. Hagan and C. C. Kao, J. Mol. Biol., 2012, 419, 284–300 CrossRef CAS PubMed.
- C. Carrasco, A. Luque, M. Hernando-Perez, R. Miranda, J. L. Carrascosa, P. A. Serena, M. d. Ridder, A. Raman, J. Gomez-Herrero, I. A. T. Schaap, D. Reguera and P. J. d. Pablo, Biophys. J., 2011, 100, 1100–1108 CrossRef CAS PubMed.
- C. Carrasco, A. Carreira, I. A. T. Schaap, P. A. Serena, J. Gomez-Herrero, M. G. Mateu and P. J. d. Pablo, PNAS, 2006, 103, 13706–13711 CrossRef CAS PubMed.
- R. Vaughan, B. Tragesser, P. Ni, X. Ma, B. Dragnea and C. C. Kao, J. Virol., 2014, 88, 6483–6491 CrossRef CAS PubMed.
- J. Helenius, C.-P. Heisenberg, H. E. Gaub and D. J. Muller, J. Cell Sci., 2008, 121, 1785–1791 CrossRef CAS PubMed.
- E. Lamers, J. t. Riet, M. Domanski, R. Luttge, C. G. Figdor, J. G. E. Gardeniers, X. F. Walboomers and J. A. Jansen, Eur. Cells Mater., 2012, 23, 182–194 CAS.
- T. Bornschlögl, S. p. Romero, C. L. Vestergaard, J.-F. o. Joanny, G. T. V. Nhieu and P. Bassereau, PNAS, 2013, 110, 18928–18933 CrossRef PubMed.
- G. Zeng, T. Müller and R. L. Meyer, Langmuir, 2014, 30, 4019–4025 CrossRef CAS PubMed.
- C.-C. Wu, D. N. Reinhoudt, C. Otto, V. Subramaniam and a. A. H. Velders, Small, 2011, 7, 989–1002 CrossRef CAS PubMed.
- G. Arrabito, S. Reisewitz, L. Dehmelt, P. I. Bastiaens, B. Pignataro, H. Schroeder and C. M. Niemeyer, Small, 2013, 9, 4243–4249 CrossRef CAS PubMed.
- J. E. Bear and J. M. Haugh, Curr. Opin. Cell Biol., 2014, 30, 74–82 CrossRef CAS PubMed.
- L. S. Wong, C. V. Karthikeyan, D. J. Eichelsdoerfer, J. Micklefield and C. A. Mirkin, Nanoscale, 2012, 4, 659–666 RSC.
- N. P. Westcott, W. Luo and M. Yousaf, J. Colloid Interface Sci., 2014, 43, 207–213 CrossRef PubMed.
- A. Boisen, S. Dohn, S. S. Keller, S. Schmid and M. Tenje, Rep. Prog. Phys., 2011, 74, 036101–036131 CrossRef.
- C. Steffens, F. L. Leite, C. C. Bueno, A. Manzoli and P. S. D. P. Herrmann, Sensors, 2012, 12, 8278–8300 CrossRef CAS PubMed.
- S. Hsieh, S.-L. Hsieh, C.-w. Hsieh, P.-C. Lin and C.-H. Wu, J. Anal. Methods Chem., 2013, 2013, 1–5 CrossRef PubMed.
- S. Aghayee, C. Benadiba, J. Notz, S. Kasas and D. a. G. Longo, J. Mol. Recognit., 2013, 26, 590–595 CrossRef CAS PubMed.
- N. Jung, H. Seo, D. Lee, C. Y. Ryu and S. Jeon, Macromolecules, 2008, 41, 6873–6875 CrossRef CAS.
- H. C. McCaig, E. Myers, N. S. Lewis and M. L. Roukes, Nano Lett., 2014, 14, 3728–3732 CrossRef CAS PubMed.
- K. S. Hwang, S.-M. Lee, S. K. Kim, J. H. Lee and a. T. S. Kim, Annu. Rev. Anal. Chem., 2009, 2, 77–98 CrossRef CAS PubMed.
- C. J. Fee, Methods Mol. Biol., 2013, 996, 287–312 CAS.
- C. J. Fee, Methods Mol. Biol., 2013, 996, 313–322 CAS.
- N. Kodera, D. Yamamoto, R. Ishikawa and T. Ando, Nature, 2010, 468, 72–77 CrossRef CAS PubMed.
- I. Chourpa, F. H. Lei, P. Dubois, M. Manfait and G. D. Sockalingum, Chem. Soc. Rev., 2008, 37, 993–1000 RSC.
- T. Schmid, L. Opilik, C. Blum and R. Zenobi, Angew. Chem., Int. Ed., 2013, 52, 5940–5954 CrossRef CAS PubMed.
- C. Blum, T. Schmid, L. Opilik, S. Weidmann, S. R. Fagerer and R. Zenobi, J. Raman Spectrosc., 2012, 43, 1895–1904 CrossRef CAS.
- B. Kainz, E. A. Oprzeska-Zingrebe and J. L. Toca-Herrera, Biotechnol. J., 2014, 9, 51–60 CrossRef CAS PubMed.
- B. Harke, J. V. Chacko, H. Haschke, C. Canale and A. Diaspro, Opt. Nanosc., 2012, 1, 1–6 CrossRef PubMed.
- K.-A. D. Walker, S. H. Doak and P. R. Dunstan, Ultramicroscopy, 2011, 111, 1200–1205 CrossRef CAS PubMed.
- K.-A. D. Walker, C. Morgan, S. H. Doak and P. R. Dunstan, PLoS One, 2012, 7, e31592 CAS.
- H. A. Huckabay, K. P. Armendariz, W. H. Newhart, S. M. Wildgen and R. C. Dunn, in Nanoimaging: methods and protocols, ed. A. A. Sousa and M. J. Kruhlak, Springer, 2013 Search PubMed.
- P. Hinterdorfer, M. F. Garcia-Parajo and Y. F. Dufrene, Acc. Chem. Res., 2012, 45, 327–336 CrossRef CAS PubMed.
- K. Klein, A. M. Gigler, T. Aschenbrenner, R. Monetti, W. Bunk, F. Jamitzky, G. Morfill, R. W. Stark and J. Schlegel, Biophys. J., 2012, 102, 360–368 CrossRef CAS PubMed.
- M. Knauer, N. P. Ivleva, X. Liu, R. Niessner and C. Haisch, Anal. Chem., 2010, 82, 2766–2772 CrossRef CAS PubMed.
- M. Fries and A. Steele, in Confocal Raman Microscopy, ed. T. Dieing, O. Hollricher and J. Toporski, Springer, 2010 Search PubMed.
- K. Wormuth, in Confocal Raman Microscopy, ed. T. Dieing, O. Hollricher and J. Toporski, Springer, 2010 Search PubMed.
- U. Schmidt, J. Müller and J. Koenen, in Confocal Raman Microscopy, ed. T. Dieing, O. Hollricher and J. Toporski, Springer, 2010 Search PubMed.
- T. Wermelinger and R. Spolenak, in Confocal Raman Microscopy, ed. T. Dieing, O. Hollricher and J. Toporski, Springer, 2010 Search PubMed.
- R. Treffer, R. Bohme, T. Deckert-Gaudig, K. Lau, S. Tiede, X. Lin and V. Deckert, Biochem. Soc. Trans., 2012, 40, 609–614 CrossRef CAS PubMed.
- S. Najjar, D. Talaga, L. Schue, Y. Coffinier, S. Szunerits, R. Boukherroub, L. Servant, V. Rodriguez and S. Bonhommeau, J. Phys. Chem. C, 2014, 118, 1174–1181 CAS.
- A. Tarun, N. Hayazawa and S. Kawata, Anal. Bioanal. Chem., 2009, 394, 1775–1785 CrossRef CAS PubMed.
- E. A. Pozzi, M. D. Sonntag, N. Jiang, J. M. Klingsporn, M. C. Hersam and R. P. V. Duyne, ACS Nano, 2013, 7, 885–888 CrossRef CAS PubMed.
- T. Deckert-Gaudig and V. Deckert, Curr. Opin. Chem. Biol., 2011, 15, 719–724 CrossRef CAS PubMed.
- K. F. Domke and B. Pettinger, ChemPhysChem, 2010, 11, 1365–1373 CrossRef CAS PubMed.
- A. P. D. Elfick, A. R. Downes and R. Mouras, Anal. Bioanal. Chem., 2010, 396, 45–52 CrossRef CAS PubMed.
| This journal is © the Owner Societies 2015 |