Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Monovalent maleimide functionalization of gold nanoparticles via copper-free click chemistry†
D. J.
Nieves
a,
N. S.
Azmi
ab,
R.
Xu
ac,
R.
Lévy
a,
E. A.
Yates
a and
D. G.
Fernig
*a
aInstitute of Integrative Biology, University of Liverpool, Crown Street, Liverpool, UK L69 7ZB. E-mail: dgfernig@liv.ac.uk
bFaculty of Industrial Sciences and Technology, Universiti Malaysia Pahang, Kuantan, Malaysia
cCentre for Molecular Oncology, Barts Cancer Institute, Queen Mary, University of London, London, EC1M 6BQ
First published on 9th September 2014
Abstract
A single maleimide was installed onto the self-assembled monolayer of gold nanoparticles by copper-free click chemistry. Simple covalent biofunctionalisation is demonstrated by coupling fibroblast growth factor 2 and an oligosaccharide in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry by thiol-Michael addition.
1 stoichiometry by thiol-Michael addition.
Gold nanoparticles have been used in biology for several decades as a contrast agent in immunoelectron microscopy. In these early applications, antibodies were adsorbed to the nanoparticle surface.1,2 More recently, advances in passivation of nanoparticle surfaces with small hydrophilic ligands have provided the means to conjugate proteins and peptides to nanoparticles stoichiometrically.3–6 Allied to new techniques in optical microscopy,7–9 highly sensitive analysis of biological interactions in living cells has been achieved, providing new insights into biological processes.5,10,11
An important challenge for single molecule microscopies is control over the stoichiometry of the biomolecule to the nanoparticle. Lack of such control can confound or alter the observed biological behaviour due to multivalency12 or exchange.13 Stoichiometric coupling of proteins to gold nanoparticles has been achieved through genetically encoded affinity tags,3,6 electrostatic charge14 and biomolecular recognition,15 among others.16 These approaches, although leading to a stable conjugate, are non-covalent. Thus, there remains the concern that the conjugated protein will exchange and, therefore, alter the biological functionality of the nanoparticle.13 Moreover, only some proteins and biomolecules can be conjugated in this way, as others, e.g., polysaccharides and nucleic acids, may require a quite different conjugation route. Therefore, an approach that allowed a chemically versatile stoichiometric covalent linkage to be formed between biological molecule and nanoparticle, in a structurally defined position, would be of great interest.
Covalent linkage between nanoparticles and biomolecules has been demonstrated via diverse chemical routes including those that are bioorthogonal (i.e., chemistry that can occur within biological settings without interfering with endogenous biochemistry). One such approach is the functionalisation of nanomaterials with azide groups that serve as a platform for reactions with strained cyclooctynes via both copper-catalysed and copper-free click chemistry.17–20 Particularly in the cases of copper-free methods, this was essential to avoid the unfavourable consequences a copper catalyst can have on biological systems through Fenton reactions.21,22 However, although some control over the number of reactive groups present has been demonstrated,20 monovalency has not. As monovalency is especially pertinent in the fields of single molecule biophysics and biology, then a route to provide control over number of labels per biomolecule is highly desired.3,12,23
A maleimide is an attractive functional group for the covalent labelling of biomolecules, due to its stability in water (cf., N-hydroxysuccinimide, commonly used to react with amines). The maleimide has a double bond, which will react readily with thiols to form a covalent thioether linkage, known as thiol-Michael addition.24,25 Thiols are present on the side chain of cysteine residues in proteins, but are relatively scarce. Moreover, the thiol group of cysteine, unless involved in a catalytic site, can often be replaced by the hydroxyl group of serine, with no discernable biological consequences, e.g., in FGF2.26 This allows a single free thiol to be engineered in a protein at a defined site, e.g., N-terminus. Thiols may also be installed specifically on nucleic acids and polysaccharides.27,28 Thus, a maleimide functionalised nanoparticle is potentially a very versatile tool.
Here we describe the indirect installation of a maleimide group on a nanoparticle via copper-free click chemistry. Gold nanoparticles with azide function were generated by the incorporation of a small percentage of an azide functionalised alkane-thiol ethylene glycol ligand into a self-assembled monolayer (SAM) of a mixture of peptidol and alkane-thiol ethylene glycol ligands (structures of all ligands are presented in Fig. S1, ESI†). The constituents of the monolayer, referred to as a ‘mix matrix’, and the rationale for the ligand proportions have been described previously and shown to impart very high stability to the nanoparticles, including resistance to ligand exchange and to non-specific binding in living cells.3,11 Upon addition of the azide ligand, the other constituent ligands are proportionally scaled to allow for the percentage of functional ligand in the ligand mix. Hence, the ligand mix consisted of 99% (mol/mol) of 2 mM mix-matrix ligands (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, CVVVT-ol to HS-C11-EG4-OH) supplemented with 1% (mol/mol) azide ligand (HS-C11-EG5-N3). One tenth volume of phosphate-buffered saline (10× PBS: 1.4 M NaCl, 27 mM KCl, 81 mM Na2PO4, 12 mM KH2PO4, pH 7.4) supplemented with Tween-20 0.1% (v/v) was added as buffer after the addition of matrix ligands, to provide electrolytes to drive self-assembly of the monolayer, then left overnight with mixing on a rotary wheel. Excess free ligand was then removed from the nanoparticles by Sephadex G-25 size-exclusion chromatography with 1× Tris-buffered saline (20 mM Tris, 150 mM NaCl, pH 7.6) with Tween-20 0.05% (v/v) (TBST0.05%) as the mobile phase. TBS is the recommended buffer for the reaction with the dibenzocyclooctyne functionalised with a maleimide group (Click-IT® Maleimide DIBO Alkyne, Life Technologies, UK).
30, CVVVT-ol to HS-C11-EG4-OH) supplemented with 1% (mol/mol) azide ligand (HS-C11-EG5-N3). One tenth volume of phosphate-buffered saline (10× PBS: 1.4 M NaCl, 27 mM KCl, 81 mM Na2PO4, 12 mM KH2PO4, pH 7.4) supplemented with Tween-20 0.1% (v/v) was added as buffer after the addition of matrix ligands, to provide electrolytes to drive self-assembly of the monolayer, then left overnight with mixing on a rotary wheel. Excess free ligand was then removed from the nanoparticles by Sephadex G-25 size-exclusion chromatography with 1× Tris-buffered saline (20 mM Tris, 150 mM NaCl, pH 7.6) with Tween-20 0.05% (v/v) (TBST0.05%) as the mobile phase. TBS is the recommended buffer for the reaction with the dibenzocyclooctyne functionalised with a maleimide group (Click-IT® Maleimide DIBO Alkyne, Life Technologies, UK).
To evaluate the effect of the azide ligand on the resistance of the particles to ligand exchange, following size-exclusion chromatography on Sephadex G-25, the azide functionalised gold nanoparticles (N3NPs) were subjected to a DTT ligand-exchange assay, as previously described.29 The UV-vis absorption spectrum for the reaction was recorded at various time intervals and the stability parameter calculated (Fig. S2, ESI†). The stability of the nanoparticles containing azide ligand in the SAM was identical to that of nanoparticles possessing a SAM of only mix-matrix ligands. Both are resistant to ligand exchange in the presence of 10 mM DTT for up to 48 h (Fig. S2a and b, ESI†). At higher concentrations of DTT, i.e., 25 and 50 mM, nanoparticle aggregation was observed after 24 h, since the stability parameter increased, which indicated that the monolayer was undergoing ligand exchange. This is consistent with the previously reported stability to ligand exchange of nanoparticles stabilised by such SAMs.29,30
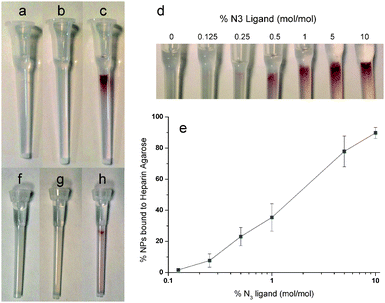
The N3NPs were then used for a copper-free click reaction with 20 μM of maleimide functionalised cyclooctyne (DIBO-Mal) for one hour in the dark with mixing on a rotating wheel to yield maleimide functionalised gold nanoparticles (Fig. S3, ESI†). After removal of excess DIBO-Mal by Sephadex G-25 size-exclusion chromatography with TBST0.05% as the mobile phase, the resulting nanoparticles (DIBO-Mal NPs) were then used for Michael addition reactions with molecules bearing thiol groups. FGF2 has previously been stoichiometrically conjugated to nanoparticles via a N-terminal hexahistidine tag and its biological activity correlated with its ability to bind to heparin.3,11,31 FGF2 possesses two surface thiols, one of which is fully exposed (Fig. S4, ESI†). FGF2 was incubated at 35 times molar excess over DIBO-Mal NPs with mixing for 3 h on a rotating wheel. The mixture was then purified by heparin agarose affinity chromatography using 1× PBS with Tween-20 0.05% (v/v) (PBST0.05%) as the mobile phase (Fig. 1). Nanoparticles bearing no maleimide functionality, i.e., a mix matrix SAM, when incubated with FGF2 did not bind to the column (Fig. 1a). This was also the case with DIBO-Mal NPs (Fig. 1b). Moreover, when the DIBO-Mal NPs were incubated with FGF2 protein that had its surface thiols blocked (by reaction with and excess of biotin-maleimide), no nanoparticles bound to the column (Fig. S5, ESI†). However, when the DIBO-Mal NPs were incubated with FGF2 protein, the pink colour of the nanoparticles was seen on the heparin agarose column (Fig. 1c). The nanoparticles were eluted with 2 M NaCl, as for native FGF2.3 Thus, the reaction of DIBO-Mal NPs with FGF2 protein only proceeded via a thiol-Michael addition route.
We have shown previously that affinity chromatography can be used to evaluate the valency of functional nanoparticles.4 Particles that have one or more FGF2 proteins can be distinguished easily from those that have none by their capacity to bind to heparin. Assuming a Poisson distribution for the number of ligands per nanoparticle, we estimated that, when 10% of the nanoparticles were bound, the nanoparticle population would possess an average of 0.1 functional ligands per nanoparticle, which corresponds to 10% of the nanoparticles possessing at least one functional ligand (with no more than 0.48% possessing two or more functional ligands) and 90% no functional ligand.3,4 To demonstrate control over the number of azide groups incorporated into the mix matrix SAM, a titration was performed varying the molar percentage of azide ligands (0–10%; mol/mol) in the ligand mix. The nanoparticles were reacted with a 35-fold molar excess of FGF2 protein for 3 h. Heparin affinity chromatography was used to determine the number of FGF2 functionalised nanoparticles, and thereby the number of azide groups reacted with DIBO-Mal per nanoparticle (Fig. 1d). The nanoparticles that do not bind, and are present in the flow through are those lacking an azide group or those that failed to react with DIBO-MAL. From the flow through the concentration of nanoparticles bound to the heparin affinity column was quantified by UV-vis spectrometry32 (Fig. 1e). The percentage of nanoparticles bound to the heparin column increased as the mol% of N3 ligand increased (Fig. 1e). No binding was observed in the absence of azide ligand (Fig. 1d). The concentration at which 10% of the nanoparticles bound to the heparin column was a molar ratio of 0.28% (mol/mol) azide ligand to matrix ligands (Fig. 1h). From this point, to prepare monovalent maleimide functionalised nanoparticles, the initial N3NPs were synthesized with 0.28% (mol/mol) azide ligand in their monolayer. It is important to note that this titration may hold for the current batches of ligands and nanoparticles. Different batches have differences in, e.g., peptidol purity, nanoparticle size, so the titration curve will be shifted.
To demonstrate the versatility of maleimide functionalised nanoparticles, advantage was taken of the resistance of the mix matrix ligand shell to DTT induced ligand exchange (Fig. S2, ESI†). This would allow the DIBO-Mal NPs to be reacted with dithiols, e.g., dithiothreitol (DTT) to yield a thiol-functionalised nanoparticle (SH NP; Fig. S6, ESI†) that can be reacted with a maleimide functionalised biological molecule. To this end we used a heparin-derived oligosaccharide (degree of polymerisation (dp)12), possessing a reducing-end maleimide function, dp12-Mal (Fig S6, ESI†). The dp12 oligosaccharide for the reaction was generated via the digestion and purification of porcine mucosal heparin (Celsus Laboratories, Cincinati, OH, USA; compound purity Table S1 and NMR characterisation Table S2, ESI†).33 A Schiff base reaction with 2 mg of freeze-dried dp12 and 100 μL of 15 mg mL−1 4-(4-N-maleimidophenyl)butyric acid hydrazide-HCl (MPBH; Thermo Fischer Scientific Inc., Rockford, IL, USA) was performed. This provided the means to couple sugars with a reducing end to the maleimide.34 The dp12-Mal product was then purified via anion-exchange chromatography on a Hi-Trap Q column (Fig. S7, ESI†). A clear peak was observed at 35 min, with absorbance at 230 nm owing to the chromophore within the MPBH structure (Fig. S7, ESI†).
To install a thiol on the surface of the nanoparticles, DIBO-Mal NPs were incubated with 1 mM DTT for 1 hour, after which excess DTT was removed via Sephadex G-25 size-exclusion chromatography with PBST0.05% as the mobile phase. The nanoparticles were then reacted with dp12-Mal. Chromatography of the mixture on an anion-exchange resin, DEAE-Sepharose, to which the dp12 binds, was used to determine the presence of oligosaccharides on the nanoparticle with PBST0.05% as the mobile phase. Nanoparticles with a non-functionalised mix monolayer incubated with oligosaccharide dp12 did not bind to the DEAE-Sepharose (Fig. 1f) and neither did SH NPs (Fig. 1g). However, when SH NPs had been incubated with the dp12 they bound to the DEAE-Sepharose, indicated by a pink colour at the top of the column (Fig. 1h). Thus, nanoparticles only bind to the resin when the Michael addition reaction has occurred between SH NPs and the maleimide modified oligosaccharides. The nanoparticles functionalised with maleimide-modified dp12 were eluted from the DEAE resin using 2 M NaCl.
In summary, we have demonstrated control over the number of maleimide groups down to monovalency into a monolayer that is resistant to ligand exchange via copper-free click chemistry. These nanoparticles provide a platform for the successful covalent conjugation of FGF2 protein, the installation of a thiol functional group on the nanoparticles and reaction with a maleimide functionalised oligosaccharide, dp12-Mal, via Michael addition reactions. With the control over the number of maleimide groups on the nanoparticle established, these nanoparticle conjugates are stoichiometrically coupled at a ratio of one nanoparticle to one biological molecule. DIBO-Mal NPs show great potential for the covalent labelling of biomolecules, or even other nanomaterials, that can undergo Michael addition reactions.
Notes and references
- R. J. Peach, W. A. Day, P. J. Ellingsen and A. R. McGiven, Histochem. J., 1988, 20, 156–164 CrossRef CAS.
- C. V. Clevenger and A. L. Epstein, Exp. Cell Res., 1984, 151, 194–207 CrossRef CAS.
- L. Duchesne, D. Gentili, M. Comes-Franchini and D. G. Fernig, Langmuir, 2008, 24, 13572–13580 CrossRef CAS PubMed.
- R. Levy, Z. X. Wang, L. Duchesne, R. C. Doty, A. I. Cooper, M. Brust and D. G. Fernig, ChemBioChem, 2006, 7, 592–594 CrossRef CAS PubMed.
- Y. Suzuki, C. N. Roy, W. Promjunyakul, H. Hatakeyama, K. Gonda, J. Imamura, B. Vasudevanpillai, N. Ohuchi, M. Kanzaki, H. Higuchi and M. Kaku, Mol. Cell. Biol., 2013, 33, 3036–3049 CrossRef CAS PubMed.
- M. De, S. Rana and V. M. Rotello, Macromol. Biosci., 2009, 9, 174–178 CrossRef CAS PubMed.
- S. Berciaud, D. Lasne, G. Blab, L. Cognet and B. Lounis, Phys. Rev. B: Condens. Matter Mater. Phys., 2006, 73 Search PubMed.
- K. Murase, T. Fujiwara, Y. Umemura, K. Suzuki, R. Iino, H. Yamashita, M. Saito, H. Murakoshi, K. Ritchie and A. Kusumi, Biophys. J., 2004, 86, 4075–4093 CrossRef CAS PubMed.
- Y. Sako and A. Kusumi, J. Cell Biol., 1994, 125, 1251–1264 CrossRef CAS.
- C. Leduc, S. Si, J. Gautier, M. Soto-Ribeiro, B. Wehrle-Haller, A. Gautreau, G. Giannone, L. Cognet and B. Lounis, Nano Lett., 2013, 13, 1489–1494 CAS.
- L. Duchesne, V. Octeau, R. N. Bearon, A. Beckett, I. A. Prior, B. Lounis and D. G. Fernig, PLoS Biol., 2012, 10, e1001361 CAS.
- A. Saha, S. Basiruddin, A. R. Maity and N. R. Jana, Langmuir, 2013, 29, 13917–13924 CrossRef CAS PubMed.
- S. Mori, H. K. Takahashi, K. Yamaoka, M. Okamoto and M. Nishibori, Life Sci., 2003, 73, 93–102 CrossRef CAS.
- R. Hong, N. O. Fischer, A. Verma, C. M. Goodman, T. Emrick and V. M. Rotello, J. Am. Chem. Soc., 2004, 126, 739–743 CrossRef CAS PubMed.
- M. Zheng and X. Huang, J. Am. Chem. Soc., 2004, 126, 12047–12054 CrossRef CAS PubMed.
- S. Rana, Y. C. Yeh and V. M. Rotello, Curr. Opin. Chem. Biol., 2010, 14, 828–834 CrossRef CAS PubMed.
- J. L. Brennan, N. S. Hatzakis, T. R. Tshikhudo, N. Dirvianskyte, V. Razumas, S. Patkar, J. Vind, A. Svendsen, R. J. M. Nolte, A. E. Rowan and M. Brust, Bioconjugate Chem., 2006, 17, 1373–1375 CrossRef CAS PubMed.
- A. Bernardin, A. Cazet, L. Guyon, P. Delannoy, F. Vinet, D. Bonnaffe and I. Texier, Bioconjugate Chem., 2010, 21, 583–588 CrossRef CAS PubMed.
- P. Gobbo, S. Novoa, M. C. Biesinger and M. S. Workentin, Chem. Commun., 2013, 49, 3982–3984 RSC.
- E. Oh, K. Susumu, J. B. Blanco-Canosa, I. L. Medintz, P. E. Dawson and H. Mattoussi, Small, 2010, 6, 1273–1278 CrossRef CAS PubMed.
- R. V. Lloyd, P. M. Hanna and R. P. Mason, Free Radical Biol. Med., 1997, 22, 885–888 CrossRef CAS.
- D. R. Lloyd and D. H. Phillips, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1999, 424, 23–36 CrossRef CAS.
- J. Imamura, Y. Suzuki, K. Gonda, C. N. Roy, H. Gatanaga, N. Ohuchi and H. Higuchi, J. Biol. Chem., 2011, 286, 10581–10592 CrossRef CAS PubMed.
- J. D. Gregory, J. Am. Chem. Soc., 1955, 77, 3922–3923 CrossRef CAS.
- D. P. Nair, M. Podgórski, S. Chatani, T. Gong, W. Xi, C. R. Fenoli and C. N. Bowman, Chem. Mater., 2013, 26, 724–744 CrossRef.
- M. Seno, R. Sasada, M. Iwane, K. Sudo, T. Kurokawa, K. Ito and K. Igarashi, Biochem. Biophys. Res. Commun., 1988, 151, 701–708 CrossRef CAS.
- R. Karamanska, B. Mukhopadhyay, D. A. Russell and R. A. Field, Chem. Commun., 2005, 3334–3336, 10.1039/b503843j.
- S. S. Ghosh, P. M. Kao, A. W. McCue and H. L. Chappelle, Bioconjugate Chem., 1990, 1, 71–76 CrossRef CAS.
- X. Y. Chen, W. W. Qoutah, P. Free, J. Hobley, D. G. Fernig and D. Paramelle, Aust. J. Chem., 2012, 65, 266–274 CrossRef CAS.
- F. Schulz, T. Vossmeyer, N. G. Bastus and H. Weller, Langmuir, 2013, 29, 9897–9908 CrossRef CAS PubMed.
- A. Yayon, M. Klagsbrun, J. D. Esko, P. Leder and D. M. Ornitz, Cell, 1991, 64, 841–848 CrossRef CAS.
- W. Haiss, N. T. Thanh, J. Aveyard and D. G. Fernig, Anal. Chem., 2007, 79, 4215–4221 CrossRef CAS PubMed.
- M. A. Skidmore, A. F. Dumax-Vorzet, S. E. Guimond, T. R. Rudd, E. A. Edwards, J. E. Turnbull, A. G. Craig and E. A. Yates, J. Med. Chem., 2008, 51, 1453–1458 CrossRef CAS PubMed.
- S. M. Chamow, T. P. Kogan, D. H. Peers, R. C. Hastings, R. A. Byrn and A. Ashkenazi, J. Biol. Chem., 1992, 267, 15916–15922 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Thiolated ligand structures, characterisation of N3NPs resistance to ligand exchange, annotated crystal structure of FGF2 protein, chromatographic characterisation of maleimide sugar starting material and product, 1H and 13C NMR of heparin starting material. See DOI: 10.1039/c4cc05909c |
| This journal is © The Royal Society of Chemistry 2014 |