Open Access Article
Open Access ArticleEvaluating the sensitivity of hybridization-based epigenotyping using a methyl binding domain protein†
Brandon W.
Heimer
a,
Tatyana A.
Shatova
a,
Jungkyu K.
Lee
b,
Kaja
Kaastrup
a and
Hadley D.
Sikes
*a
aDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02129, USA. E-mail: sikes@mit.edu
bDepartment of Chemistry, Kyungpook National University, Daegu, South Korea
First published on 28th April 2014
Abstract
Hypermethylation of CpG islands in gene promoter regions has been shown to be a predictive biomarker for certain diseases. Most current methods for methylation profiling are not well-suited for clinical analysis. Here, we report the development of an inexpensive device and an epigenotyping assay with a format conducive to multiplexed analysis.
Introduction
Accurately diagnosing and characterizing cancer is critical to improving patient outcomes. Medical research coupled with analytical tests has started to allow physicians to identify cohorts of patients and deliver therapy developed for their particular needs. For example, markers such as BRCA1 and HER2 are routinely used for screening, prognosis, or treatment selection.1 In addition to somatic mutations, research over the past decade has shown that epigenetic disruptions in gene expression, particularly tumor suppressors, can lead to neoplastic disease.2 Current indications show it is likely the FDA will soon evaluate DNA methylation profiles, specifically in the context of the genes PITX2 (ref. 3) (breast cancer) and SEPT9 (ref. 4) (colon cancer), for use as cancer biomarkers because such diagnostics have the potential to provide additional information to that available from the current methods used in the clinic.In mammals, enzymatic methylation of the cytosine base occurs only in the context of the dinucleotide sequence CpG, that is, when cytosine is sequentially followed by guanosine on the same DNA strand.2 The human genome has short stretches of DNA with a high density of CpG dinucleotides termed “CpG islands.” Many CpG islands are coincident with gene promoters.5 These CpGs are usually unmethylated.6 However, in colorectal cancer, human mutL homolog 1 (hMLH1) promoter hypermethylation has been shown to contribute to genetic hypermutability as a consequence of inactivated DNA mismatch repair function.7 Others have correlated promoter hypermethylation to patients' responsiveness to select chemotherapeutics. For example, a subset of glioblastoma patients with silenced O6-methylguanine DNA methyltransferase (MGMT) is particularly responsive to treatment with alkylating agents because there is no active enzyme to remove alkyl adducts from DNA.8 CpG island hypermethylation has further been shown to be unique to the tissue of origin9 which adds to the promise of DNA hypermethylation profiles as cancer biomarkers.
Methylation-specific PCR10 is the de facto standard for methylation profiling in diagnostic settings where the genes of interest are known. Methylation-specific PCR, however, requires sodium bisulfite conversion of unmethylated cytosine bases to urasil in the target DNA. Chemical conversion is costly, and the reaction alone requires 12–16 h. Methods must be assiduously optimized to minimize incomplete deamination of unmethylated cytosine bases and inappropriate conversion of methylated ones to thymine; both errors can lead to inaccurate results.11 The degenerate genomic sequence following conversion can also constrain the primer sequence used for downstream PCR amplification.12 Further, readout is not direct. The converted DNA must be amplified using a separate PCR reaction for each CpG interrogated and analyzed by gel electrophoresis making it unsuitable for multiplexing. Luo et al. proposed an alternative in which a methyl binding domain (MBD) protein facilitates detection.13 However, this format necessitates not only the bisulfite conversion and PCR steps required for methylation-specific PCR but also enzymatic remethylation of the DNA. Recently, Yu et al. reported using surface plasmon resonance (SPR) to detect MBD binding to methylated DNA.14 While requiring fewer steps, this method is also less amenable to multiplexed detection and the reliance on ProteOn™ GLC Sensor Chips (BioRad) significantly increases the cost per test.
Here, we report the development of a simple method for detecting methylated DNA fragments from the MGMT gene promoter. A defining feature is that target oligonucleotides from the test sample hybridize directly to capture probes printed in 300 μm diameter spots on an inexpensive biochip without requiring bisulfite conversion. We detect methylated DNA duplexes using an MBD protein engineered to facilitate detection using either fluorescence or photopolymerization-based signal amplification with short reaction times.15
Results and discussion
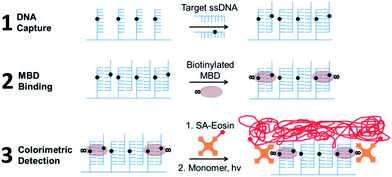
Our epigenotyping assay follows the scheme shown in Fig. 1. First, single-stranded, 5′-hydrazide modified DNA capture probes are spotted onto an aldehyde functional biochip according to the layout illustrated in Fig. 2a. Any remaining unreacted aldehyde groups are quenched with bovine serum albumin (BSA). DNA target oligonucleotides in a test sample then hybridize to the corresponding sequence-specific capture probes on the biochip. Next, biotinylated MBD proteins bind to DNA duplexes with symmetrically methylated CpGs. Streptavidin functionalized with either eosin (photoinitiator) or Cy3 dye then transduces MBD binding into a colorimetric or fluorescent readout as depicted in Fig. 2b. For Cy3 labeling, the biochip is directly scanned to produce the type of image shown in Fig. 2c. For colorimetric detection, an aqueous polyethylene glycol diacrylate monomer is polymerized to form hydrogels specifically at the sites with surface-bound eosin when irradiated with green (522 nm) light for approximately two minutes.15 The hydrogels become visible to the unaided eye after staining with a red dye for two minutes as shown in Fig. 2d.15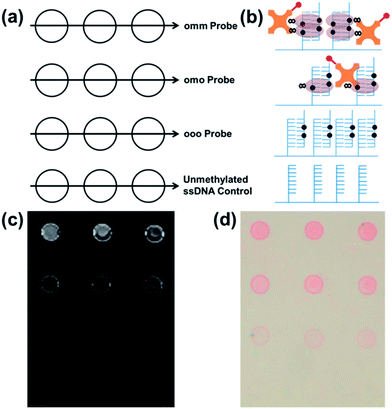 | ||
| Fig. 2 (a) Biochips were spotted with capture probe ssDNA oligos having two (omm), one (omo), or no (ooo) methylated CpGs in order to epigenotype the target oligos. Unmethylated ssDNA served as the negative control. (b) Schematic representation of the area within each group of spots following hybridization with 100 nM doubly methylated (omm) target ssDNA and detection using the scheme shown in Fig. 1. Fluorescence (c) and colorimetric (d) readout of MBD binding to methylated DNA. | ||
Several design choices were key to obtaining the results presented in Fig. 2. The epigenotyping assay we describe here uses a standard glass microscope slide coated with an agarose film.20 The agarose film coating, with its 3D surface structure, shields biomolecules from the charged glass substrate and provides a solution-like environment conducive for nucleic acid and protein binding events.22,25 Experimentally, the agarose film was essential to our assay because self-assembled monolayer (SAM) coated glass surfaces, when evaluated, suffered from high nonspecific binding and signals not distinguishable from background; see ESI Fig. S4.† Agarose film coated surfaces also benefit from being significantly less expensive than SAMs. The capture probe oligos were modified with a hydrazide which has a significantly higher reactivity toward aldehyde functional surfaces than primary amines leading to detectable DNA hybridization at sub-nanomolar concentrations as a result of the higher probe density;26,27 see ESI Fig. S5.† Each DNA probe was spotted at 10 μM to saturate the aldehyde functional surface with capture probe oligonucleotides. Further, we sought to print the smallest spots (300 μm diameter) that allowed for unaided, colorimetric readout; such feature size miniaturization has also been shown to improve surface capture performance.28 The average surface density of the biotinylated control oligo was measured to be 900 ± 40 oligos per μm2. When the un- (ooo), singly- (omo), and doubly-methylated (omm) capture probes were hybridized with 100 nM complimentary, biotinylated target oligos, the average surface density of the duplex decreased slightly to 850 ± 50 duplex per μm2; see ESI Fig. S6† for further detail. All further binding signals were reported as a fraction of the capture probe density, assumed equal to the measured density of biotinylated ssDNA (900 ± 40 oligos per μm2).
The MBD protein is expressed as a green fluorescent protein (GFP) fusion. In addition to being a fluorescent reporter, the GFP also significantly improves the soluble yield of the fusion when induced using the protocol we optimized previously.18 We further added a 15 amino acid C-terminal AviTag™ (Avidity) with an internal lysine residue which the co-expressed BirA biotin ligase enzyme biotinylates. Such in vivo biotinylation greatly simplifies MBD production and eliminates the risk of denaturation and loss of binding activity following chemical biotinylation which requires organic solvents. The MBD concentration used in the assay was tuned to maximize signal from duplex DNA with both singly, and doubly, symmetrically methylated CpGs and minimize signal from hemi-methylated (defined as signal from unmethylated (ooo) probe spots hybridized with methylated (omo or omm) target oligos) and background signal (defined as signal from unmethylated ssDNA). Using a MBD concentration of 40 μg mL−1 optimized signal intensity, specificity, and the amount of MBD protein used per assay (1.6 μg) as shown in ESI Fig. S7;† thus, it was used for all further experiments. Maintaining a low background signal is critical for achieving low fluorescence signal-to-noise ratios (SNR). However, the ratio is less critical for the colorimetric assay because no hydrogels will be formed as long as the background signal is below the photopolymerization amplification threshold.22
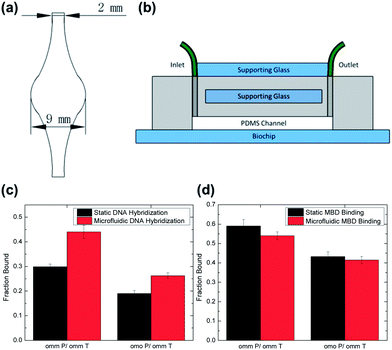
Nucleic acid hybridization is typically performed under inefficient, diffusion-limited conditions in conventional microarray experiments.29 Numerous microfluidic devices that use mixing have been reported to improve DNA hybridization.29–31 We fabricated a reusable microfluidic device from PDMS, shown in Fig. 3a, to implement a recirculating mixing method similar to that described by Lee et al. for the purpose of improving DNA hybridization as well as standardize the work flow and provide an assay format suitable for automation. Consistent with previous reports, hybridizing target DNA using the microfluidic device produced MBD binding signals approximately one-third higher than those achieved using static DNA incubation over the times studied; see Fig. 3c.30 However, experimental results shown in Fig. 3d indicated no similar improvement in the fraction bound using the flow system for MBD binding.
In order to more completely understand whether convection, diffusion and/or reaction rate were limiting the MBD binding step for the purpose of improving assay sensitivity, we calculated both the Peclet and the Damköhler numbers for the flow system.23,24 The Peclet number is a measure of the importance of convection relative to diffusion. It was found to be 1700 which implies the presence of a thin (approximately 7 μm) depletion region of MBD above the binding surface. Any MBD above the depletion region is swept down the channel without having a chance to encounter the methylated DNA. This also means the mass transport toward the binding surface is only weakly dependent on the flow rate of the system. Furthermore, the Damköhler number, which is a ratio of the intrinsic rate of reaction to diffusion, was calculated to be 0.3 using the depletion region length-scale. Because this value is less but not much less than unity, the system is only partially reaction-limited. The predicted weak dependence of MBD binding on flow rate and the partial reaction limit both seem to explain the equivalent performance of both systems for the MBD binding step.
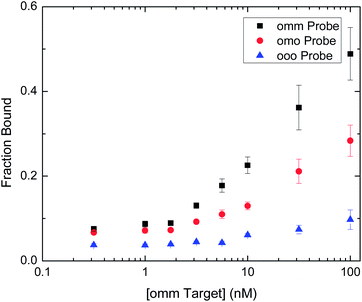
The signal from MBD binding versus the concentration of target ssDNA with two methylated CpG sites (omm) is shown in Fig. 4 for capture probes with two, one or no complimentary, methylated CpG sites. We determined the limit of quantification for MBD binding to spots with two or one symmetrically methylated CpG sites using our epigenotyping assay to be 0.3 nM and 1 nM, respectively, as defined by the target concentration that produces a signal greater than that from the un-methylated probe plus ten standard deviations.32 Sample size is the current drawback of the system as the reported sensitivity requires a larger number of cells than what can be obtained from a needle biopsy. In a clinical sample, the two MGMT alleles may be heterozygously methylationed at a CpG dinucleotide in the interrogated promoter sequence. All cells in the population would, however, share this methylation pattern due to the clonal expansion of tumor cells and heritability of DNA methylation.33 Therefore, the LOD curve would reside between the unmethylated (ooo) and singly methylated (omo) curves in that case.
Conclusions
We have demonstrated a novel method for directly hybridizing and detecting methylated DNA fragments on a biochip. This simple method requires less time to perform than the existing methods which rely on bisulfite conversion of unmethylated cytosine bases, PCR, and/or next-generation DNA sequencing. Eliminating sodium bisulfite treatment further reduces the risk of DNA degradation and inaccuracies resulting from incomplete conversion. Our method is also able to achieve a low cost per test by using inexpensive test surfaces and a recombinant MBD protein that can be produced efficiently in E. coli. Fluorescence detection requires a microarray scanner for readout; however, the colorimetric assay requires only a green light source and digital camera for imaging the visible, stained hydrogels.New biochips can easily be customized to interrogate a specific gene or panel of genes by simply designing unique capture probes specific to the sequences of interest. Further, microfluidic integration provides continuous delivery of test solutions, standardizes the assay, and provides a platform well-suited for automation. Quantitative experimental and theoretical analysis also enables future work to develop technologies using engineered MBD proteins for single-cell epigenetic analyses.
Experimental
MBD protein cloning, expression, purification, and characterization
The biotin acceptor peptide sequence MAGGLNDIFEAQKIEWHE was appended to the C-terminal end of the murine MBD1-green fluorescent protein (MBD-GFP) construct14 from Steve Blair's Lab (University of Utah, Salt Lake City) for in vivo biotinylation by the biotin ligase enzyme BirA. This was accomplished using PCR-driven overlap extension. In short, we PCR amplified the MBD-GFP gene from the pET-30b vector starting 4 bp upstream of the EcoRI restriction site and ending immediately before the 3′ TAA stop codon following the GFP cDNA. Fwd primer: 5′-ATCCGAATTCGATGCCAAAAAAGA-3′ (24 nt, GC = 37.5%, Tm = 55.6 °C) rev. primer: 5′-CTTGTACAGCTCGTCCATGC-3′ (20 nt, GC = 55.0%, Tm = 56.0 °C) concurrently, we synthesized the cDNA for the biotin acceptor sequence from primers designed using the DNAWorks software from the NIH.16 The construct began at the 5′ end with a 30 bp sequence complimentary to the end of the GFP cDNA (ATCACTCCCGGCATGGACGAGCTGTACAAG) followed by the cDNA for the acceptor sequence, the ochre stop codon, a one nucleotide spacer “A”, the terminal XhoI restriction site and a 4 nt 3′ overhang to facilitate restriction enzyme binding. The MBD1-GFP PCR product and biotin acceptor construct were joined using PCR-driven overlap extension.17Fwd primer: 5′-ATCCGAATTCGATGCCAAAAAAGA-3′ (24 nt, GC = 37.5%, Tm = 55.6 °C) rev. primer: 5′-GGTGCTCGAGTTTATTCATGCCATTCAATTTTCTGCG-3′ (37 nt, GC = 43.2%, Tm = 63.5 °C).
The gene structure is shown in ESI Fig. S1.† The pET-30b vector and PCR product were double digested with EcoRI-HF and XhoI (New England BioLabs), gel purified, and ligated using T4 ligase (New England BioLabs). The pET-30b/MBD1-GFP-B construct was co-transformed with pACYC184/BirA (Avidity) into chemically competent E. coli BL21 (DE3) cells (New England BioLabs).
E. coli BL21 (DE3) cells containing both plasmids were inoculated from a glycerol cell stock into a 14 mL culture tube (VWR Scientific) containing 5 mL of Difco LB Broth (BD Biosciences) supplemented with 10 μg mL−1 chloramphenicol (Calbiochem) and 50 μg mL−1 kanamycin (Calbiochem) and grown overnight at 37 °C and 250 rpm orbital shaking. These cells were then subcultured in a 1 L flask containing 250 mL of TB medium (12 g tryptone (BD Biosciences), 24 g yeast extract (BD Biosciences), and 4 mL glycerol (BDH Chemicals) dissolved in 900 mL of 18 MΩ deionized (DI) water, autoclaved, cooled, and brought to 1 L with the addition of 100 mL of a sterile solution of 0.17 M KH2PO4 (Macron Chemicals) and 0.72 M K2HPO4 (BDH Chemicals) supplemented with 50 μg mL−1 kanamycin and incubated at 37 °C and 250 rpm orbital shaking. When the expression culture reached an OD600 of 0.6, MBD expression was induced by adding IPTG (Omega Bio-Tek) and D-Biotin (Sigma-Aldrich) to final concentrations of 1 mM and 50 μM, respectively, and decreasing the incubator temperature to 20 °C for 16 h.18 Cells were harvested in five 50 mL conical tubes (BD Falcon) by centrifuging at 4000g and 4 °C for 10 min.
One tube was subsequently lysed with BugBuster HT Protein Extraction Reagent (EMD Millipore) following the manufacturer's instructions. The soluble fraction was separated from insoluble cell debris by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min. The clarified lysate was filtered using Acrodisc PF Syringe Filter 0.8/0.2 (Pall) and loaded onto a 1 mL HisTrap™ Fast Flow column (GE Healthcare) for purification on an ÄKTA purifier 10™ FPLC System (GE Healthcare). The column was equilibrated and washed with binding buffer19 (300 mM NaCl (Mallinckrodt Chemicals), 100 mM NaxHxPO4 (Mallinckrodt Chemicals), 10 mM Tris–HCl (BDH Chemicals), 10 mM 2-mercaptoethanol (J. T. Baker), and 10 mM imidazole (Alfa Aesar), pH 8), and MBD protein was eluted using a linear gradient from 0% to 100% elution buffer19 (300 mM NaCl, 100 mM NaxHxPO4, 10 mM Tris–HCl, 10 mM 2-mercaptoethanol, and 250 mM imidazole, pH 8) over 20 column volumes (CV) followed by a 5 CV isocratic elution at 100% elution buffer. The pH of the binding and elution buffers was raised from 7.4 to 8 to eliminate a co-eluting impurity present in our prior expression and purification work with MBD-GFP fusion proteins.18 The fractions containing only MBD protein were pooled, concentrated, and buffer exchanged into storage buffer (1× PBS (AMRESCO), 10% glycerol and 1 mM dithiothreitol (MP Biomedicals)) using a 30
000g for 15 min. The clarified lysate was filtered using Acrodisc PF Syringe Filter 0.8/0.2 (Pall) and loaded onto a 1 mL HisTrap™ Fast Flow column (GE Healthcare) for purification on an ÄKTA purifier 10™ FPLC System (GE Healthcare). The column was equilibrated and washed with binding buffer19 (300 mM NaCl (Mallinckrodt Chemicals), 100 mM NaxHxPO4 (Mallinckrodt Chemicals), 10 mM Tris–HCl (BDH Chemicals), 10 mM 2-mercaptoethanol (J. T. Baker), and 10 mM imidazole (Alfa Aesar), pH 8), and MBD protein was eluted using a linear gradient from 0% to 100% elution buffer19 (300 mM NaCl, 100 mM NaxHxPO4, 10 mM Tris–HCl, 10 mM 2-mercaptoethanol, and 250 mM imidazole, pH 8) over 20 column volumes (CV) followed by a 5 CV isocratic elution at 100% elution buffer. The pH of the binding and elution buffers was raised from 7.4 to 8 to eliminate a co-eluting impurity present in our prior expression and purification work with MBD-GFP fusion proteins.18 The fractions containing only MBD protein were pooled, concentrated, and buffer exchanged into storage buffer (1× PBS (AMRESCO), 10% glycerol and 1 mM dithiothreitol (MP Biomedicals)) using a 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MWCO spin filter (Millipore). MBD concentration was assayed using the bicinchoninc acid (BCA) protein assay kit – reducing agent compatible (Thermo Scientific) with BSA standards. Approximately 1.5 μg of MBD protein was loaded onto a 4–15% SDS-PAGE gradient gel (Bio-Rad) and visualized with Coomassie Brilliant Blue R250 stain (AMRESCO); see ESI Fig. S2a.† MBD binding activity to methylated DNA was verified by electromobility shift assay (EMSA) using the method described previously;18 see ESI Fig. S2b.† Purified protein was stored at −80 °C in 20 μL single-use aliquots for later use in methylation profiling assays.
000 MWCO spin filter (Millipore). MBD concentration was assayed using the bicinchoninc acid (BCA) protein assay kit – reducing agent compatible (Thermo Scientific) with BSA standards. Approximately 1.5 μg of MBD protein was loaded onto a 4–15% SDS-PAGE gradient gel (Bio-Rad) and visualized with Coomassie Brilliant Blue R250 stain (AMRESCO); see ESI Fig. S2a.† MBD binding activity to methylated DNA was verified by electromobility shift assay (EMSA) using the method described previously;18 see ESI Fig. S2b.† Purified protein was stored at −80 °C in 20 μL single-use aliquots for later use in methylation profiling assays.
Streptavidin mutein matrix (Roche) was used for the final polishing affinity chromatography step to remove any MBD protein not biotinylated in vivo. Roche's batch purification protocol and 50 μL of resin were used to purify 125 μg of monobiotinylated MBD after already being purified using IMAC as described above. Three elution fractions were collected and pooled. An UltraCruz™ Micro G-25 (Santa Cruz Biotechnology) gel filtration spin column was used to remove free biotin remaining from eluting the biotinylated MBD protein. Pure MBD was buffer exchanged into storage buffer as described above. The final polishing purification step was not routinely performed after it was found not to increase the signal from MBD binding to methylated DNA in the biochip assay; see ESI Fig. S3.†
Preparing biochip test surfaces
Standard clear glass microscope slides (VWR Scientific) coated with an aldehyde functionalized agarose film were prepared using 2 mL of a 0.2% w/v SeaKem® LE agarose (Lonza) solution and the method previously described.20 The concentration of agarose in the film casting solution was decreased to 0.2% w/v from 1% reported by Afanassiev et al. to reduce nonspecific uptake of the eosin Y stain used to visualize the photopolymerized hydrogels formed usain PBA detection. The ooo, omo, and omm probe oligonucleotides previously designed to capture target DNA sequences from the MGMT promoter14 were ordered from Integrated DNA Technologies with a 5′ hydrazide functionalization (instead of biotin used by Yu et al.) to facilitate coupling to the reactive aldehyde groups on the biochip surface. Additionally, two, 50 nt single-stranded oligonucleotides (5′-CATCACACAACATCACACAACATCACGTATATAAAACGGAACGTCGAAGG-3′) were designed as controls; both had 5′ hydrazide functionalization and one had a biotin covalently attached to the 3′ end. Each probe was diluted to 10 μM in 3× SSC buffer (G Biosciences) and spotted at 50% relative humidity according to the layout illustrated in Fig. 2a using a GeSiM Nano-Plotter 2.1 equipped with a Nano Tip. Each spot was formed from ten separate 300 pL depositions. After spotting, each biochip was transferred to a vacuum desiccator for overnight incubation. A circular, 9 mm diameter isolator well was cut from Scotch 3M 665 tape and affixed to the biochip to define each test area. Each biochip was then rinsed under a stream of DI water and blown dry using compressed nitrogen gas. Biochips ready for testing were stored in the vacuum desiccator until needed.Microfluidic device fabrication
Microfluidic devices were fabricated using photolithography techniques.21 A 4′′ silicon wafer was coated with a 60 μm thick layer of SU-8 2050 (MicroChem) and patterned using a 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 dpi transparency mask (CAD/Art Services, Inc.). Next, a thin layer of PDMS (Sylgard 184, Dow Corning) was cast onto the SU-8 mold and cured for 15 min at 80 °C until solid. Then, a supporting glass piece was cut from a standard microscope slide (VWR Scientific) to be 20 mm × 12 mm, and was positioned over the microfluidic channel. The rest of the PDMS was poured onto the mold and cured for 2 h at 80 °C. The PDMS was then peeled off and each device was cut out separately. The inlet and outlet holes were punched using a 1.5 mm ID × 1.91 mm OD Harris Uni-Core Puncher (Ted Pella, Inc.). The microfluidic channel, shown in Fig. 3a, features 2 mm wide inlet and outlet regions that expand to a 9 mm diameter circular test area. The channel was designed to be 27 mm long in order to allow for a standard glass slide (25 mm wide) to be used as an additional support above the PDMS device; see Fig. 3b. The device was then positioned above the test area on the biochip slide and clamped on using standard binder clips (Staples).
400 dpi transparency mask (CAD/Art Services, Inc.). Next, a thin layer of PDMS (Sylgard 184, Dow Corning) was cast onto the SU-8 mold and cured for 15 min at 80 °C until solid. Then, a supporting glass piece was cut from a standard microscope slide (VWR Scientific) to be 20 mm × 12 mm, and was positioned over the microfluidic channel. The rest of the PDMS was poured onto the mold and cured for 2 h at 80 °C. The PDMS was then peeled off and each device was cut out separately. The inlet and outlet holes were punched using a 1.5 mm ID × 1.91 mm OD Harris Uni-Core Puncher (Ted Pella, Inc.). The microfluidic channel, shown in Fig. 3a, features 2 mm wide inlet and outlet regions that expand to a 9 mm diameter circular test area. The channel was designed to be 27 mm long in order to allow for a standard glass slide (25 mm wide) to be used as an additional support above the PDMS device; see Fig. 3b. The device was then positioned above the test area on the biochip slide and clamped on using standard binder clips (Staples).
Detecting methylated DNA with MBD, fluorescence and PBA
To block each test area against nonspecific protein binding, each isolator well was filled with 40 μL of 1% w/v BSA in 1× PBS and incubated for 15 min in a humid chamber at ambient temperature. Excess BSA was then washed away with 1× PBS, and the entire biochip was submersed in 1× PBS at 4 °C for 10 min. Following a rinse with DI water, dilutions of omm target14 oligonucleotides were prepared in hybridization buffer (6× SSC, 5× Denhardt's (BioExpress)), and 40 μL of each were pipetted onto separate test areas. Biochips were then incubated in a humid chamber (defined here as a pipette tip box partially filled with DI water) for 4 h at ambient temperature to allow for DNA capture. Each biochip was rinsed with 1× PBS followed by DI water to remove unhybridized target DNA. Next, 40 μL of MBD protein diluted in binding buffer (20 mM HEPES (EMD Millipore), pH 7.9, 3 mM MgCl2 (BDH Chemicals), 10% v/v glycerol, 1 mM dithiothreitol, 100 mM KCl (BDH Chemicals), 0.1% w/v BSA, and 0.01% v/v Tween-20) was pipetted onto each test area, and the biochip was incubated for 30 min in a humid chamber at 4 °C. After MBD incubation, each test site was rinsed with PBST (1× PBS, 0.1% Tween-20 (Sigma-Aldrich)) followed by 1× PBS, and the entire biochip was submersed in 1× PBS at 4 °C for 25 min. Following a rinse with DI water, 40 μL of 0.1 μM streptavidin-Cy3 or streptavidin-eosin in 1× PBS, 5× Denhardt's, and 0.5% w/v BSA was pipetted onto each test area, and the biochip was incubated for 5 min in a humid chamber covered in foil at ambient temperature. Unbound labeling reagent was removed by washing sequentially with PBST, 1× PBS, and DI water followed by blow drying using compressed nitrogen gas. The biochip was either scanned in the case of streptavidin-Cy3 fluorescent detection or polymerized using an aqueous PEG diacrylate monomer and methods previously described.22Alternatively, the reusable microfluidic device was attached to a rinsed and dried biochip using binder clips. FEP 1/16′′ OD × 0.020′′ ID tubing (Upchurch Scientific) was inserted into each port on the device and mated to 0.89 mm ID Tygon-LFL tubing (Saint-Gobain) which was then run through a Masterflex C/L 77120-52 peristaltic pump (Cole Parmer). All of the tubing connections were self-sealing, and no glue was necessary. A 1% w/v BSA in 1× PBS solution was pumped through the microfluidic device at 100 μL min−1 for 15 min followed by 1× PBS for 10 min. A solution of omm target ssDNA in 6× SSC/5× Denhardt's was recirculated at 100 μL min−1 for 4 h. A 1× PBS solution was pumped through the microfluidic device at 100 μL min−1 for 5 min followed by DI water for another 5 min. MBD protein diluted to 40 μg mL−1 in binding buffer was recirculated at 100 μL min−1 for 30 min. PBST, 1× PBS, and DI water were sequentially pumped through the microfluidic device for 5 min each at 100 μL min−1. Fluorescent or photoinitiator labeling and detection was performed as described above.
Biochip analysis
Biochips labeled with streptavidin-Cy3 were scanned using a Molecular Devices GenePix 4000B fluorescent microarray scanner. Each fluorescence image was analyzed using ImageJ (NIH). The mean fluorescence intensity for each spot was determined by adjusting the threshold of the image to include the entire spot area and averaging the constituent pixel intensities. Fraction bound values were computed by dividing the average signal from replicate spots by the average signal from the biotinylated ssDNA control spots. The background corrected intensity (BCI) was computed by subtracting the mean background intensity (signal from unmethylated ssDNA) from the mean fluorescence intensity value from replicate spots. Cy3 surface densities were then calculated from BCI values using a calibration curve generated by scanning and analyzing a Full Moon Biosystems calibration array. Dividing the Cy3 surface densities by the known average number of Cy3 dye molecules per streptavidin produced the surface density of binding events.Each polymerized test area was imaged using the digital camera in the ampliPHOX® Reader (InDevR) imaging bay.
Quantitative MBD hybridization analysis
The MBD diffusion coefficient (D) was estimated to be 1.1 × 10−10 m2 (s)−1 using the Stokes–Einstein equation. All calculations used 2.88 × 106 (M s)−1 and 0.4 (s)−1 MBD association (ka) and dissociation (kd) rates, respectively.14 The depletion zone thickness was approximated as Ld = L(Pes)−1/3, where Pes is defined as the shear Peclet number (Pes = 6(L/H)2Pe).23,24L is the length of the binding area, which was approximated as 900 μm (equivalent to three spot diameters), H is the channel height, Pe is the Peclet number (Pe = UH/D), and U is the fluid velocity. The Damköhler number (Da = kaCsLd/D) was calculated based on the surface density of DNA duplexes (Cs) in each spot and the depletion zone thickness (Ld).Acknowledgements
A David H. Koch (1962) first-year graduate fellowship to BWH, a NSF Graduate Research Fellowship to KK, a Burroughs Wellcome Fund Career Award at the Scientific Interface to HDS, the MIT Center for Environmental Health Sciences (through support from the NIH-NIEHS Center Grant P30-ES002109), the James H. Ferry Fund for Innovation, and the Joseph R. Mares endowed chair supported this work. We thank Prof. Steve Blair for the gift of the pET-30b+/MBD-GFP plasmid, Prof. Allan S. Myerson for the use of his GeSiM Nano-Plotter, the staff of the MIT Microsystems Technology Laboratories for technical support, and InDevR, Inc. for the gift of two ampliPHOX® Readers. We thank Felice Frankel for help with the design of Figure 1 and for the photograph in Figure 2D.Notes and references
- J. A. Ludwig and J. N. Weinstein, Nat. Rev. Cancer, 2005, 5, 845–856 CrossRef CAS PubMed.
- J. G. Herman and S. B. Baylin, N. Engl. J. Med., 2003, 349, 2042–2054 CrossRef CAS PubMed.
- O. Hartmann, F. Spyratos, N. Harbeck, D. Dietrich, A. Fassbender, M. Schmitt, S. Eppenberger-Castori, V. Vuaroqueaux, F. Lerebours, K. Welzel, S. Maier, A. Plum, S. Niemann, J. A. Foekens, R. Lesche and J. W. M. Martens, Clin. Cancer Res., 2009, 15, 315–323 CrossRef CAS PubMed.
- T. deVos, R. Tetzner, F. Model, G. Weiss, M. Schuster, J. Distler, K. V. Steiger, R. Grützmann, C. Pilarsky, J. K. Habermann, P. R. Fleshner, B. M. Oubre, R. Day, A. Z. Sledziewski and C. Lofton-Day, Clin. Chem., 2009, 55, 1337–1346 CAS.
- D. Macleod, R. R. Ali and A. Bird, Mol. Cell. Biol., 1998, 18, 4433–4443 CAS.
- A. P. Bird, Nature, 1986, 321, 209–213 CrossRef CAS PubMed.
- J. G. Herman, A. Umar, K. Polyak, J. R. Graff, N. Ahuja, J.-P. J. Issa, S. Markowitz, J. K. V. Willson, S. R. Hamilton, K. W. Kinzler, M. F. Kane, R. D. Kolodner, B. Vogelstein, T. A. Kunkel and S. B. Baylin, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6870–6875 CrossRef CAS.
- M. E. Hegi, A.-C. Diserens, T. Gorlia, M.-F. Hamou, N. de Tribolet, M. Weller, J. M. Kros, J. A. Hainfellner, W. Mason, L. Mariani, J. E. C. Bromberg, P. Hau, R. O. Mirimanoff, J. G. Cairncross, R. C. Janzer and R. Stupp, N. Engl. J. Med., 2005, 352, 997–1003 CrossRef CAS PubMed.
- J. F. Costello, M. C. Fruhwald, D. J. Smiraglia, L. J. Rush, G. P. Robertson, X. Gao, F. A. Wright, J. D. Feramisco, P. Peltomaki, C. James, D. E. Schuller, L. Yu, C. D. Bloomfield, M. A. Caligiuri, A. Yates, R. Nishikawa, H.-J. Su Huang, N. J. Petrelli, X. Zhang, M. S. O'Dorisio, W. A. Held, W. K. Cavenee and C. Plass, Nat. Genet., 2000, 24, 132–138 CrossRef CAS PubMed.
- J. G. Herman, J. R. Graff, S. Myöhänen, B. D. Nelkin and S. B. Baylin, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 9821–9826 CrossRef CAS.
- D. P. Genereux, W. C. Johnson, A. F. Burden, R. Stöger and C. D. Laird, Nucleic Acids Res., 2008, 36, e150 CrossRef PubMed.
- S. J. Clark, A. Statham, C. Stirzaker, P. L. Molloy and M. Frommer, Nat. Protoc., 2006, 1, 2353–2364 CrossRef CAS PubMed.
- J. Luo, W. Zheng, Y. Wang, Z. Wu, Y. Bai and Z. Lu, Anal. Biochem., 2009, 387, 143–149 CrossRef CAS PubMed.
- Y. Yu, S. Blair, D. Gillespie, R. Jensen, D. Myszka, A. H. Badran, I. Ghosh and A. Chagovetz, Anal. Chem., 2010, 82, 5012–5019 CrossRef CAS PubMed.
- K. Kaastrup and H. D. Sikes, Lab Chip, 2012, 12, 4055–4058 RSC.
- D. M. Hoover and J. Lubkowski, Nucleic Acids Res., 2002, 30, e43 CrossRef PubMed.
- K. L. Heckman and L. R. Pease, Nat. Protoc., 2007, 2, 924–932 CrossRef CAS PubMed.
- M. E. Boyd, B. W. Heimer and H. D. Sikes, Protein Expression Purif., 2012, 82, 332–338 CrossRef CAS PubMed.
- H. F. Jørgensen, K. Adie, P. Chaubert and A. P. Bird, Nucleic Acids Res., 2006, 34, e96 CrossRef PubMed.
- V. Afanassiev, V. Hanemann and S. Wölfl, Nucleic Acids Res., 2000, 28, e66 CrossRef CAS PubMed.
- D. C. Duffy, J. C. McDonald, O. J. A. Schueller and G. M. Whitesides, Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS PubMed.
- K. Kaastrup, L. Chan and H. D. Sikes, Anal. Chem., 2013, 85, 8055–8060 CrossRef CAS PubMed.
- T. M. Squires, R. J. Messinger and S. R. Manalis, Nat. Biotechnol., 2008, 26, 417–426 CrossRef CAS PubMed.
- T. Gervais and K. F. Jensen, Chem. Eng. Sci., 2006, 61, 1102–1121 CrossRef CAS PubMed.
- M. Dufva, Biomol. Eng., 2005, 22, 173–184 CrossRef CAS PubMed.
- S. Raddatz, J. Mueller-Ibeler, J. Kluge, L. Wäβ, G. Burdinski, J. R. Havens, T. J. Onofrey, D. Wang and M. Schweitzer, Nucleic Acids Res., 2002, 30, 4793–4802 CrossRef CAS PubMed.
- L. M. Johnson, R. R. Hansen, M. Urban, R. D. Kuchta and C. N. Bowman, Biomacromolecules, 2010, 11, 1133–1138 CrossRef CAS PubMed.
- D. S. Dandy, P. Wu and D. W. Grainger, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 8223–8228 CrossRef CAS PubMed.
- J. Liu, B. A. Williams, R. M. Gwirtz, B. J. Wold and S. Quake, Angew. Chem., Int. Ed., 2006, 45, 3618–3623 CrossRef CAS PubMed.
- H. H. Lee, J. Smoot, Z. McMurray, D. A. Stahl and P. Yager, Lab Chip, 2006, 6, 1163–1170 RSC.
- R. H. Liu, R. Lenigk, R. L. Druyor-Sanchez, J. Yang and P. Grodzinski, Anal. Chem., 2003, 75, 1911–1917 CrossRef CAS.
- G. L. Long and J. D. Winefordner, Anal. Chem., 1983, 55, 712A–724A CrossRef CAS.
- R. Jaenisch and A. Bird, Nat. Genet., 2003, 33, 245–254 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4an00667d |
| This journal is © The Royal Society of Chemistry 2014 |