Influence of anion–water interactions on the behaviour of lipases in room temperature ionic liquids†
Muhammad Alif Mohammad Latifa,
Nuno M. Micaêloc and
Mohd Basyaruddin Abdul Rahman*ab
aDepartment of Chemistry, Faculty of Science, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor Darul Ehsan, Malaysia. E-mail: basya@upm.edu.my; Fax: +60 38943 5380; Tel: +60 38946 6798
bEnzyme and Microbial Technology Research Centre, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor Darul Ehsan, Malaysia
cDepartamento de Química, Universidade do Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: micaelo@itqb.unl.pt; Fax: +351 25 3604 382; Tel: +351 25 3604 370
First published on 12th September 2014
Abstract
In this report, molecular dynamics simulations were applied in order to investigate the effect of Room Temperature Ionic Liquid (RTIL) anions toward the structure and dynamic properties of lipases. Two lipases were studied; Candida antarctica lipase B and Candida rugosa lipase were solvated by five RTILs that contained the same cation, with increasing hydration levels. Several properties were investigated: structural deviations and flexibility of the protein conformation, the behaviour of RTILs at the protein surface, and the interactions between RTILs and water molecules in the systems. Both lipases' conformations showed an increased structural stability in RTILs when compared to an aqueous solution. The lowest structural deviation was observed around 15 to 20 percent of water content (w/w protein). The RTIL with the chloride anion was shown to be the exception however, inducing the least structural stability at low water percentages. The flexibility of both lipases was clearly affected when transferred from aqueous into RTILs. The flexible regions found for both lipases in water were significantly more rigid in RTILs. Around the protein surface, the behaviour of RTIL anions and the water molecules was similar to other conventional organic solvents. The water retention ability for all RTIL anions was consistent for both lipases except for the bis(trifluoromethylsulfonyl)imide anion, which showed distinctive behaviour toward different protein surface properties. The effect of water content was more profound compared to the difference between the RTILs anions studied. However, it was found that the structural and dynamic properties of the lipases were affected by the behaviour of anions toward the hydration layer of the enzymes.
Introduction
In the field of biocatalysis, it has been well established that Room Temperature Ionic Liquids (RTIL)s can influence enzyme activity and reaction rate when used as non-aqueous media or as enzyme-supports.1,2 RTILs, which are composed entirely of cations and anions, offer mild reaction conditions in addition to increase in yields.3,4 The majority of enzymes that were reported to be active in RTILs belong to the class of lipases.5 Lipases such as Candida rugosa lipase (CRL) and Candida antarctica lipase B (CALB), have been extensively studied in organic media and are among the most frequent enzymes studied in RTILs.6–10 For years researchers have been trying to systematically quantify the contributions of different RTIL's cation–anion combinations toward enzyme's stability and activity. Lue, et al.11 studied the enzymatic acylation reactions of flavonoids with long chain fatty acids (palmitic, oleic acids), which were carried out in 14 different RTIL media containing a range of cation and anion structures. They found that anion selection had a far greater influence on the lipase activity than the choice of the cationic moiety. RTILs containing [Tf2N]−, [PF6]− and [BF4]− anions were determined as successful reaction media while RTILs that contain anions with stronger solvating properties such as hydrogen-bonding ability, resulted in a decreased in yield. It has been reported that the physicochemical properties of cations were dependent on chain length.12,13 The same cannot be said of anions, as most of the common ones have a distinctive structural property. A [PF6]− anion may just possess two more fluorine atoms than [BF4]−, but the physicochemical properties of RTIL of these anions such as density, viscosity, hydrophobicity and polarity to name a few, are significantly different.14,15 Interestingly, some contrary reports have been published on how lipases reacted toward different types of RTIL anions. The work from Madeira Lau and co-workers9 demonstrated the potential use of RTILs for enzyme catalysis and explained their use with lipases. They compared the reactivity of CALB in RTILs, which were [BMIM][PF6] and [BMIM][BF4], with conventional organic solvents. In all cases the reaction rates were similar for the reactions investigated. However, Vidya and Chadha16 showed that lipase favoured hydrophobic anions such as [PF6]− and [Tf2N]− more than the hydrophilic anions such as [BF4]− in their transesterification study. Lee et al.17 investigated the initial rates of transesterification for three different enzymes (CALB, Rhizomucor miehei lipase and CRL) in different RTILs. At similar water activity, they observed that the initial rates were affected by the type of anion, following a decreasing order of [Tf2N]− > [PF6]− > [TfO]− > [BF4]− ∼ [SbF6]−. They showed that the highly nucleophilic [TfO]− and [BF4]− anions contributed to a lower activity of enzyme. Hernández-Fernández et al.18 observed that the stability of CALB varied in different RTILs in the order of increasing stability: ([HMIM][PF6] > [HMIM][Tf2N] > [HMIM][BF4]). They also concluded that the enzyme's stability decreased with the increasing order of nucleophilicity. Irimescu and Kato19 observed that the rate of CALB-catalyzed enantioselective acylation of 1-phenylethylamine with 4-pentenoic acid in different RTILs followed a decreasing order of [TfO]− > [BF4]− > [PF6]− with the same cations. However, they found that [PF6]-based RTILs produced the fastest reaction rate, followed by [TfO] and [BF4]-based RTILs in acylation reaction of 2-phenyl-1-propylamine with 4-pentenoic acid. They suggested that the effects may be caused by other factors such as hydrophobicity, viscosity and impurity. Based on these reports, enzymes were found active in several RTILs containing anions such as [PF6]−, [BF4]−, [TfO]− and [Tf2N]−. It seems that the difference in enzyme's behaviour in RTILs containing anions with distinctive physicochemical properties is largely dependent on the enzymes themselves, and these differences were enlarged by other factors such as substrates and enzyme-modifications.20Investigations at the atomic level may further explain how enzymes behave in different types of RTIL anions. By using molecular modelling techniques such as molecular dynamics (MD) simulations, it is possible to provide useful information about the structural and dynamic properties of biomolecules21–24 including lipases in organic media.25,26 Previously, there have been several reports on computational studies of enzyme properties in RTILs.27–36 The earliest MD simulation study on the structure and dynamics of a protein in RTILs was reported by Micaêlo and Soares28 where the stability of cutinase in [BMIM][PF6] and [BMIM][NO3] with varying water content at temperatures of 298 K and at 343 K was investigated. The stability of cutinase in RTILs was progressing in a bell-shaped profile across the water percentages. [BMIM][PF6] was found to render native-like structure while [BMIM][NO3] destabilized the protein conformation. Klähn et al.29 reported a MD simulation study on the stability of CALB in eight different RTILs. The stability of CALB was primarily affected by the types of anions following an order of (increasing stability) [NO3]− ≪ [BF4]− < [PF6]−. The MD simulation work by Burney and Pfaendtner have already compared the properties of enzymes when solvated in between RTILs and conventional organic solvents.30 They found that CRL has the least structural deviation in [BMIM][PF6] while the has the most in water. It was also shown that the interactions on the protein surface were dominated by the RTIL anions. Most of these computational studies involved RTIL anions such as [PF6]−, [BF4]− and [NO3]−. Consistent findings from these reports showed that [NO3]− anion destabilizes enzymes. However, to our knowledge, there has been little effort on the more complex RTIL anions such as [TfO]− and [Tf2N]−. As described previously, these anions were also heavily involved in most of the experimental studies regarding enzyme activity. Therefore, a theoretical study involving these anions is important in order to properly explain the contradiction of results from experimental findings. Previously, we have reported the structural and dynamics properties of an α-chymotrypsin in [BMIM]-based RTILs with different anions such as [PF6]−, [BF4]−, [Cl]−, [TfO]− and [Tf2N]− using MD simulations at different water percentages.31 The [Cl]− anion was chosen for its physical properties while the rest of the anions were chosen because enzymes, as pointed out before, were active in them. Several properties which correlated with experimental evidences were explained. In this report, we attempt to extend our understanding by using the same RTILs towards lipases, namely CALB and CRL. We focus our study on comparing the structural deviations and the flexibility between both lipases, while also monitoring the behaviour of water and RTILs at the enzyme surface. Based on the results from our previous work, the lipases studied could exhibit different properties when simulated in different RTILs at different hydration states.28,31,37–39 As we highlighted previously from experimental reports, a certain order can be established between different anions. We anticipated that the order of anions studied will be affected by the concentration of water in the system. Based on the experimental results, we hypothesized that [TfO]− and [Tf2N]− anions will behave differently towards different enzymes and that this contributes toward the different order of activity when different enzymes were solvated in RTIL with different anions.
Methodology
System setup
Initially, two lipases (CALB and CRL) were simulated in aqueous and then transferred into five [BMIM]-based RTILs of different water content. These RTILs were [BMIM][PF6], [BMIM][BF4], [BMIM][Cl], [BMIM][TfO] and [BMIM][Tf2N]. The crystal structure for both CRL (PDB ID: 1CRL)40 and CALB (PDB ID: 1TCA)41 were obtained from the Protein Data Bank (http://www.rcsb.org). All n-acetylglucosamine atoms were removed from the crystal structures leaving only the protein and crystallographic water molecules. The initial size of the unit cell was expanded by 2.4 nm on each axis following a cubic unit representation. OPLS force-field was applied for all simulations42 while a 4-site, TIP4P model was used to represent the water molecules.43 For RTILs, an already developed and tested set of OPLS force field parameters for RTILs were employed.44–46 Each lipase was simulated in RTILs at 5, 10, 15, 20, and 50 percent of water. Tables 1 and 2 display the number of water, RTILs cation and anion molecules in more detail. For simulation in RTILs, the number of water molecules for each water percentage was calculated based on the weight of water over weight of protein (w/w protein). Using the final trajectory from MD simulations in aqueous, water molecules that were closest to the protein surface were selected according to the percentages mentioned previously.| Enzyme | %Water (w/w protein) | ||||
|---|---|---|---|---|---|
| 5 | 10 | 15 | 20 | 50 | |
| CALB | 92 | 184 | 275 | 367 | 917 |
| CRL | 158 | 317 | 475 | 634 | 1584 |
| RTIL | %Water (w/w protein) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 10 | 15 | 20 | 50 | ||||||
| Ncat | Nan | Ncat | Nan | Ncat | Nan | Ncat | Nan | Ncat | Nan | |
| (a) CALB | ||||||||||
| [BMIM][PF6] | 1410 | 1409 | 1404 | 1403 | 1398 | 1397 | 1393 | 1392 | 1358 | 1357 |
| [BMIM][BF4] | 1538 | 1537 | 1530 | 1529 | 1523 | 1522 | 1516 | 1515 | 1472 | 1471 |
| [BMIM][Cl] | 1990 | 1989 | 1980 | 1979 | 1971 | 1970 | 1961 | 1960 | 1904 | 1903 |
| [BMIM][TfO] | 1395 | 1394 | 1389 | 1388 | 1383 | 1382 | 1378 | 1377 | 1343 | 1342 |
| [BMIM][Tf2N] | 1089 | 1088 | 1085 | 1084 | 1082 | 1081 | 1078 | 1077 | 1054 | 1053 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||||
| (b) CRL | ||||||||||
| [BMIM][PF6] | 2577 | 2560 | 2567 | 2550 | 2557 | 2540 | 2547 | 2530 | 2487 | 2470 |
| [BMIM][BF4] | 2812 | 2795 | 2799 | 2782 | 2787 | 2770 | 2774 | 2757 | 2698 | 2681 |
| [BMIM][Cl] | 3277 | 3260 | 3261 | 3244 | 3244 | 3227 | 3228 | 3211 | 3130 | 3113 |
| [BMIM][TfO] | 2550 | 2533 | 2540 | 2523 | 2530 | 2513 | 2520 | 2503 | 2461 | 2444 |
| [BMIM][Tf2N] | 1991 | 1974 | 1984 | 1967 | 1978 | 1961 | 1971 | 1954 | 1930 | 1913 |
RTIL molecules were introduced into the system as the outermost layer of the system, filling up the simulation box. Packing of molecules was accomplished using the Packmol program.47 The net charge in the pure aqueous system was kept neutral by replacing water molecules with sodium or chloride counter ions. For systems in RTILs, the net charge was neutralized by varying the number of cation and anion in the starting configuration. The protonation states for titratable amino acid residues were taken at pH 7.0 for aqueous MD simulations and were unchanged during transfer to RTILs systems.48
Simulation details
All MD simulations were performed using the GROMACS v4.5 simulation package.49 Equation of motion was integrated at 2 fs intervals. Periodic boundary conditions were applied in all directions. All bonds were harmonically constrained using the LINCS algorithm.50 The energies, forces, velocities and trajectories were recorded every 500 steps. The neighbouring atoms were identified using the grid search method and the list was updated every 5 steps (10 fs). All electrostatic and van der Waals interactions were calculated explicitly up to 1.2 nm. The Particle-Mesh Ewald (PME) algorithm51 was applied for the corrections of long range electrostatic interactions. PME was applied by using the fourth order interpolation with a Fourier-spacing of 0.12 nm. All systems were kept at room temperature by the velocity-rescaling thermostat52 with a temperature coupling constant of 0.1 ps (0.01 ps for pre-equilibration). RTILs, protein, and water molecules were individually coupled in separate heat baths to ensure all components in the system were at the correct temperatures. The Berendsen's barostat was used to keep the pressure at 1.0 atm with a relaxation time of 1.5 ps (0.5 ps for aqueous).53 The isothermal compressibility was set at 4.6 × 10−5 bar−1 for aqueous systems and 4.23 × 10−5 bar−1 for systems in RTILs.54In an effort to overcome the “slow dynamics” of RTIL molecules and to achieve faster convergence, all systems were simulated using positional-restraints with a force constant of 1 × 106 kJ mol−1 nm2.28 Several steps of energy minimizations were employed for all systems. Firstly, 5000 steps of steepest-descents energy minimization were performed on the system where all heavy atoms were position-restrained. The RTIL cations and anions, water molecules, protein heavy atoms and protein Cα atoms were released from positional-restraints consecutively in further energy minimization steps. The final step of energy minimization was another 5000 steps of steepest-descents without any position-restraints condition applied. Similar steps were applied for canonical ensemble (NVT), position-restrained MD simulations, each for 500 ps (100 ps for aqueous). When necessary, extra simulations were carried out to ensure the convergence of temperature and pressure for each step of the canonical ensemble simulations. For analysis purposes, all systems were further simulated for 10 ns (6 ns for aqueous) in an isothermal–isobaric condition (NPT). All trajectory analysis on the protein structure and dynamics were performed over the last 2 ns of MD simulations. For statistical accuracy, all systems were simulated in triplicates using different initial velocities for the first NVT simulations.
Results and discussion
Structural deviations
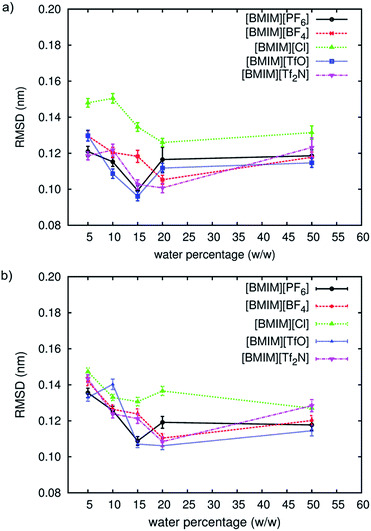
The conformational stability of CALB and CRL in aqueous and five [BMIM]-based RTILs were measured in terms of their atomic displacements, relative to the respective crystal structure. The results were represented by the average root mean square deviation (RMSD) of the protein structure, which were averaged from the last 2 ns of NPT simulations (see ESI†).The average RMSD for both lipases in RTILs at different solvation conditions is shown in Fig. 1. The first striking observation is that the lipases RMSD in all these conditions were lower than that in aqueous condition, which was 0.15 ± 0.004 nm for CALB and 0.18 ± 0.009 nm for CRL. It was clear that these enzymes were closer to their respective crystal structures in RTILs than in aqueous solution. Considering the data from aqueous simulation, the RMSD profiles for both lipases showed a bell-shaped progression across the water percentages, with an observed minimum around 10 to 20% of water. [BMIM][TfO] showed the lowest average RMSD at this percentage content, closely similar to [BMIM][PF6] and followed by [BMIM][Tf2N], [BMIM][BF4] and [BMIM][Cl]. This behaviour was similar to that found in other RTIL-solvated systems and conventional organic solvents that have been previously reported.28,31,38,39,55,56 At 15% of water, both lipases showed an order of RTIL anions (in increasing order of average RMSD) of [TfO]− ∼ [PF6]− > [Tf2N]− > [BF4]− > [Cl]−. This order agreed with the order of stability as reported by Klähn et al.29 As pointed in Fig. 1, [BMIM][Cl] demonstrated highest RMSD profiles for both enzymes when compared with other RTILs. However, the difference of average RMSD between anions, especially [Cl]− was reduced at high water percentage (50%). Similarly to what was observed in our previous study,31 the [Cl]− anions have a tendency to penetrate into the protein-core, and the presence of ions inside the protein-core have been reported to destabilize enzymes.29
This effect, however, was reduced at higher water content due to the thicker layer of water molecules surrounding the protein conformation. Other than [BMIM][Cl], average RMSD for both lipases in RTILs did not show a significant gap across the water percentages, suggesting that the structural deviations of the proteins studied here was not greatly affected by these four types of anions, but rather by the water concentration in the system. These findings also support the observation that partitioning of the hydration level between the enzyme and the bulk solvent was a determinant factor that differentiates molecular-level details in different organic media.57
From the bell-shaped RMSD profiles, it can be deduced that instead of RTIL anions, the water content (which is related to the water activity) played a major role in determining the lipases' stability in RTILs. The dependency of lipase activity on the water content have been observed experimentally, with CALB producing better initial rates when compared with CRL.17 The same report also stated that when [BMIM]+ cation was used, the initial reaction rate was hugely dependent on the nature of the anions, which is in line with the observations in this work at low water content. Schőfer et al.58 also revealed that using CALB resulted in better activity than CRL for the kinetic resolution of 1-pheenylethanol in RTILs. From these reports, CALB was shown better in RTILs when compared to CRL. However, according to Kaar et al.,59 the catalytic activity of CRL in RTILs was still better than organic solvents. They found that in [BMIM][PF6], CRL can increase the initial reaction rate up to 1.5 times faster than in hexane. In relation to the simulation results, the structural deviations of both lipases were almost identical, with only small differences found in the range of the RMSD values for each RTIL. A non-distinguished order of stability between the types of RTIL anions for these lipases was also an indication of why no definite order could be established from various experimental evidences. Therefore, it can be suggested that the difference between the lipases' activity in RTILs were not directly related to their structural stability when solvated by different RTIL anions.
Enzyme flexibility
The average root mean square fluctuations (RMSF) of the protein's main chain (Fig. 2) indicated that both enzymes were less flexible in RTILs when compared with an aqueous environment. The lowest atomic fluctuations profile was observed for CALB in [BMIM][TfO] and for CRL in [BMIM][Cl]. As observed previously in Fig. 1, hydration level can be decisive in affecting structural properties for both lipases in RTILs. At very low water content (5% w/w protein), only a small variation was observed between different RTILs. As the hydration level increased, the difference became apparent and at 50% of water (w/w protein), we could see that the average atomic fluctuations increased faster in [BMIM][PF6] and [BMIM][Tf2N] than other RTILs studied. The b-factor per residue of protein's main chain at 15% of water for both enzymes was compared with the ones in aqueous (Fig. 3). The differences between the effect of water and RTILs on both lipases' flexibility were clear in all areas but specifically distinguished at certain regions of both structures. For CALB (Fig. 3a), the flexibility as expressed by the b-factor per residue, was significantly higher in aqueous solution at the N-terminus region (residues Ser3, Gly4, Ser5, Asp6 and Pro7) while also at the very end of the amino acid sequence (residues Val315, Thr 316 and Pro317). Both of these regions were loop regions and can move easily in water. The dramatic decrease of flexibility in RTILs showed that even the freely moving loop structure of the protein lost its flexibility in RTILs. Another significant difference in flexibility between RTILs and aqueous was found between residue 146 to 148, which is a short α-helical region. Trodler and Pleiss26 previously performed short MD simulations on the structure of CALB in aqueous and non-aqueous solvents. They also found out that the flexibility as expressed by the b-factors was reduced in non-aqueous solvents, especially the hydrophobic ones.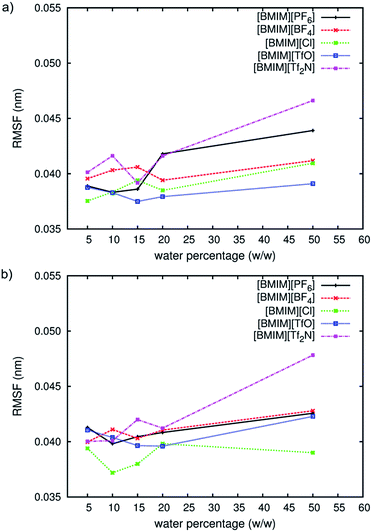 | ||
| Fig. 2 RMSF of CALB (a) and CRL (b) main chain in RTILs at different water percentages. Values are averaged from the last 2 ns of triplicate MD simulations. | ||
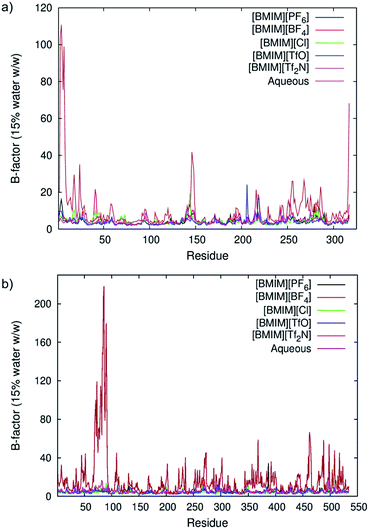 | ||
| Fig. 3 Theoretical b-factors of CALB (a) and CRL (b), calculated as per residue in aqueous and RTILs. | ||
On the other hand, CRL exhibited significantly higher flexibility in aqueous, notably from residue 69 through 91 (Tyr69, Glu70, Glu71, Asn72, Leu73, Pro74, Lys75, Ala76, Ala77, Leu78, Asp79, Leu80, Val81, Met82, Gln83, Ser84, Lys85, Val86, Phe87, Glu88, Ala89, Val90, and Ser91) which is an α-helix connected by loops at each end (Fig. 3b). These residues made up a ‘lid’ for the enzyme conformation. Tejo et al.25 also observed the lid flexibility in water in their simulation studies. They compared the flexibility of the lid in the open conformation which was used in this study (PDB ID: 1CRL) and the closed conformation (PDB ID: 1TRH). Results from their MD simulations also showed the same phenomenon where the lid of CRL for both opened and closed conformation lost its flexibility in organic solvent used (CCl4). Although enzymes were observed to be highly stable in anhydrous media, they also can produce lower activities.60 The presence of water around the surface and inside the enzyme structure is therefore important for biocatalysis as it provides flexibility toward the conformation61 and increases the enzyme-substrate interactions. The flexibility for both lipases at higher water content was clearly affected by the types of the anions. Water-immiscible anions such as [PF6]− and [Tf2N]− promoted higher flexibility while the other anions maintained a low flexibility profile across the water percentages. The decrement in flexibility caused by the presence of RTILs on both lipases studied could restrict the ability of both enzymes to catalyze substrates that are not naturally tailored to their conformations. Furthermore, the interactions of the protein surface with water and RTIL anions were also a contributing factor toward the lipases' flexibility in RTILs, which will be discussed in the following section. Meanwhile, based on our previous observation in Fig. 2, it is possible to ‘tune’ the flexibility of enzymes in RTILs by varying the water content in the system. For example, in reactions involving hydrophobic anions such as [PF6]− and [Tf2N]−, increasing the water content can contribute toward higher enzyme flexibility while not in the water-miscible anions such as [BF4]− and [TfO]−.
Surface interactions
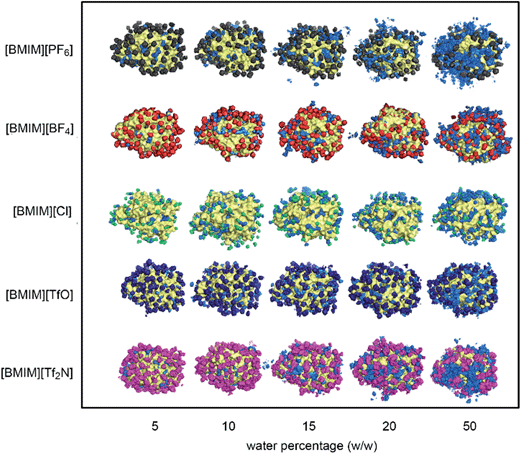
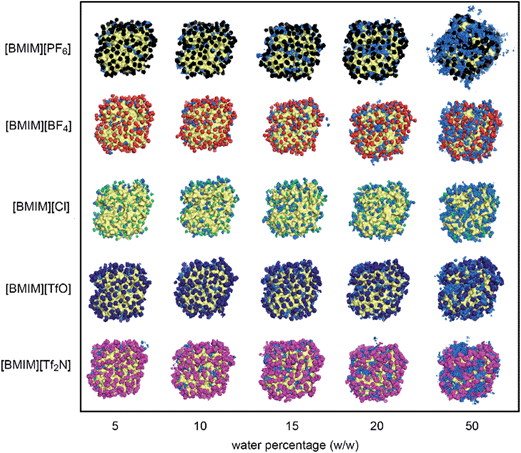
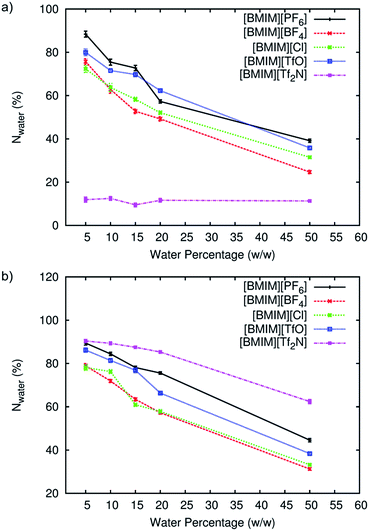
The behaviour of water and anions at the surface of the protein structure was scrutinized using spatial distributions function analysis. Water molecules were found ‘localized’ at certain regions on the protein surface, while anions formed a structured arrangement around both CALB (Fig. 4) and CRL (Fig. 5). The spaces left on the protein surface described the distributions of the cations that alternated with the anions.29 From the results, the arrangement of water and anions at protein surface was independent of the types of lipase studied. Although CRL possess a larger surface than CALB, the observed distribution of water and anions around both lipases was closely similar.In the case of [BMIM][PF6] and [BMIM][Tf2N], the anions and water molecules were highly mobile at higher water percentage, which was clear at 50%. This finding explained why the flexibility of enzyme in these RTILs increased faster at higher water content (Fig. 2). Water localization at the surface was least in [BMIM][Cl]. Due to its significantly smaller size and its high solubility in water,62 there was smaller coverage of anions around the protein surface. From this observation, it is clear that the localization of water was greatly influenced by the RTIL anions, which is further supported by the fact that localization of water molecules was favourable on the surface where the anions were covering. The interactions between RTILs and water molecules are reported to be strongly dependent on the type of anions.63,64 In this study, the behaviour of different RTIL anions and water were observed by monitoring the average percentage of water at the protein surface across the water percentages (Fig. 6). The percentage of water on the surface was determined based on the number of water located within 0.5 nm from protein surface in the starting configuration and the average number of water in the same region from the last 2 ns of MD simulations. We found that, as conventional organic solvents, RTILs stripped water molecules away from protein surface.37,64 The percentage of water at the surface for four types of anions ([PF6]−, [TfO]−, [BF4]− and [Cl]−) were consistent for both of the lipases studied. The amount of water that was stripped from the surface was increased as the water concentration increased. However, the number of water in the calculated region was actually increasing despite the strong, water-stripping capability that was exhibited by the RTILs. We estimated that the water-stripping capability of different anions was related to their tendency to hydrogen-bond with water molecules. RTILs such as [BMIM][PF6] and [BMIM][TfO] exhibited lower stripping strength due to less hydrogen bond interactions with water molecules. As for [BF4]− and [Cl]− anions, which are highly hydrophilic and therefore water-miscible,65 more water molecules were stripped away from protein surface. Previously, [BMIM][TfO] was found to reduce the enzyme flexibility but this RTIL stripped less water from the protein surface. It can be suggested that the [TfO]− anions exhibited the capability to hydrogen-bond with water through the oxygen atoms. These anions mostly interact with water at the surface as compared to the protein surface residues themselves. Hence, water could remain near the protein surface but lose their effect towards the protein flexibility.
As observed in Fig. 6a, CALB was found to maintain a certain level of hydration in [BMIM][Tf2N] despite the increasing water concentration in the system. This was also observed in our previous work.31 However, Fig. 6b showed that for CRL, the strength of water-stripping exhibited by different anions was in increasing order of [Tf2N]− < [PF6]− ∼ [TfO]− < [BF4]− ∼ [Cl]−. The variation of [BMIM][Tf2N] behaviour toward the two lipases studied can be related to the difference of the enzyme surface properties. [Tf2N]− anion is weakly-coordinated, possesses low capacity for ion-ion interactions, has poor water-solubility properties, and is most hydrophobic compared to other anions studied.66 However, the amide group in the [Tf2N]− anion is able to form strong hydrogen bond interactions with water molecules that can pull them away from the protein surface. For enzymes such as CALB and previously-studied α-Chymotrypsin,31 [BMIM][Tf2N] stripped away the most number of water molecules consistently at each water percentage studied as compared with other RTILs. However, CRL possesses more than twice the number of Asparagine residues at the surface compared to CALB (Fig. 7).
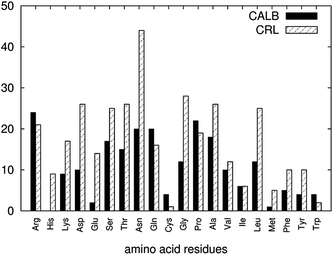 | ||
| Fig. 7 Amino acid residues that were found exposed on the surface of CALB and CRL. The amino acid residues were probed on the surface of the crystal structures for the respective enzymes. | ||
Asparagine has a high propensity to hydrogen bond, since it is possible for the amide group to accept two and donate two hydrogen bonds. The competition from these residues at the surface to hydrogen-bond with water molecules resulted in fewer [Tf2N]–water interactions. Thus, a higher percentage of water molecules was found on the surface. This finding coincided with the rapid increment of the average number of hydrogen bonds between the water and CRL in [BMIM][Tf2N] when compared to the ones in [BMIM][PF6] (Table 3). However, as the number of water found on the surface was increasing with every water percentage, the differences between the amounts of water on the surface between the RTILs did not greatly affect the overall protein–water interactions. The data from Table 3 shows that the average hydrogen-bonding between CALB in [BMIM][Tf2N] was slightly higher than [BMIM][PF6] at each water percentage. This can explain why [BMIM][Tf2N] produced a closely similar effect toward the structural deviations and flexibility of CALB, even with less water molecules on the surface when compared with other RTILs, which was consistent with our previous study.31
| %Water (w/w) | CALB | CRL | ||
|---|---|---|---|---|
| [BMIM][PF6] | [BMIM][Tf2N] | [BMIM][PF6] | [BMIM][Tf2N] | |
| 5 | 130.27 | 136.76 | 228.52 | 256.01 |
| 10 | 207.72 | 215.43 | 408.30 | 454.57 |
| 15 | 254.17 | 263.09 | 494.96 | 567.23 |
| 20 | 247.80 | 257.42 | 533.35 | 643.67 |
| 50 | 318.88 | 352.45 | 647.61 | 809.92 |
Conclusions
The structural and dynamics properties of CALB and CRL in five RTILs at different water percentages were described using MD simulations. Structural analysis indicated that both lipases were more native-like in RTILs when compared to aqueous solution. We showed that the structural properties of both enzymes were more affected by the presence of water molecules than the type of RTIL anions studied. Enzymes structural deviations showed a similar bell-shaped dependency on the water content as found in conventional organic solvents, with a minimum RMSD around 10% to 20% of water content. The size and water-solubility of RTIL anions were found to affect the stability of the protein with smaller and more water-soluble anions leading to penetration into protein-core conformation, causing instability at low water content. Although lipases were reported to be stable and active in [BMIM][BF4], our results suggested that [BMIM][PF6], [BMIM][TfO] and [BMIM][Tf2N] were more structural-friendly. We were able to show that RTILs restrained the movement of both proteins even at regions that were highly flexible in aqueous, thus affecting the selectivity of both enzymes. This is related to the grid-like arrangement of RTILs found on protein surface while water molecules were forced to localize at certain area of the proteins, consequently, reducing their effect toward enzyme flexibility. Our observations demonstrated that non-localized water molecules were stripped away from the protein surface, which behaviour is also found in conventional organic solvents. We showed that the water-stripping strength exhibited by the RTILs was related to the solubility and hydrophobicity of the anions studied. Water-immiscible anions such as [PF6]− retained more water at the surface while water-soluble anions such as [BF4]− stripped more water away from the surface. A more complex anion such as [Tf2N]− exhibited different water-stripping behaviour, which is related to the enzyme surface properties. Other RTILs showed closely similar water-stripping properties for both enzymes studied. However, as the stability and flexibility studies showed, this scenario was not influential, which signify that at low water content, [BMIM][Tf2N] is capable of providing a water-like effect to the solvated enzyme. Another factor that should be considered is the charge-interactions between the water and the RTIL anions, which can further explain their behaviour at the protein surface. Although we did not detect any major deviation toward the protein conformation, a longer MD simulation (up to microseconds) may be needed for systems such as RTILs in order to unveil other properties that were possibly isolated by this work. Advanced force fields such as the polarized ones could also be used to further distinguish the effect of using different RTIL anions towards the enzymes and water.Notes and references
- R. A. Sheldon, R. M. Lau, M. J. Sorgedrager, F. van Rantwijk and K. R. Seddon, Green Chem., 2002, 4, 147–151 RSC.
- M. B. Abdul Rahman, K. Jumbri, N. A. M. Ali Hanafiah, E. Abdulmalek, B. A. Tejo, M. Basri and A. B. Salleh, J. Mol. Catal. B: Enzym., 2012, 79, 61–65 CrossRef CAS PubMed.
- S. Park and R. J. Kazlauskas, Curr. Opin. Biotechnol., 2003, 14, 432–437 CrossRef CAS.
- Z. Yang and W. Pan, Enzyme Microb. Technol., 2005, 37, 19–28 CrossRef CAS PubMed.
- U. Kragl, M. Eckstein and N. Kaftzik, Curr. Opin. Biotechnol., 2002, 13, 565–571 CrossRef CAS.
- T. De Diego, P. Lozano, S. Gmouh, M. Vaultier and J. L. Iborra, Biomacromolecules, 2005, 6, 1457–1464 CrossRef CAS PubMed.
- S. Ha, S. Lee, D. Dang, M. Kwon, W.-J. Chang, Y. Yu, I. Byun and Y.-M. Koo, Korean J. Chem. Eng., 2008, 25, 291–294 CrossRef CAS.
- P. Lozano, T. De Diego, D. Carrié, M. Vaultier and J. L. Iborra, Biotechnol. Lett., 2001, 23, 1529–1533 CrossRef CAS.
- R. Madeira Lau, F. van Rantwijk, K. R. Seddon and R. A. Sheldon, Org. Lett., 2000, 2, 4189–4191 CrossRef CAS PubMed.
- F. van Rantwijk, F. Secundo and R. A. Sheldon, Green Chem., 2006, 8, 282–286 RSC.
- B.-M. Lue, Z. Guo and X. Xu, Process Biochem., 2010, 45, 1375–1382 CrossRef CAS PubMed.
- H. Tokuda, K. Hayamizu, K. Ishii, M. A. B. H. Susan and M. Watanabe, J. Phys. Chem. B, 2005, 109, 6103–6110 CrossRef CAS PubMed.
- D. Xiao, L. G. Hines, S. Li, R. A. Bartsch, E. L. Quitevis, O. Russina and A. Triolo, J. Phys. Chem. B, 2009, 113, 6426–6433 CrossRef CAS PubMed.
- H. Jin, B. O'Hare, J. Dong, S. Arzhantsev, G. A. Baker, J. F. Wishart, A. J. Benesi and M. Maroncelli, J. Phys. Chem. B, 2007, 112, 81–92 CrossRef PubMed.
- H. Tokuda, K. Hayamizu, K. Ishii, M. A. B. H. Susan and M. Watanabe, J. Phys. Chem. B, 2004, 108, 16593–16600 CrossRef CAS.
- P. Vidya and A. Chadha, J. Mol. Catal. B: Enzym., 2010, 65, 68–72 CrossRef CAS PubMed.
- S. Lee, Y.-M. Koo and S. Ha, Korean J. Chem. Eng., 2008, 25, 1456–1462 CrossRef CAS PubMed.
- F. J. Hernández-Fernández, A. P. de la Ríos, F. Tomás-Alonso, D. Gómez and G. Víllora, Can. J. Chem. Eng., 2009, 87, 910–914 CrossRef.
- R. Irimescu and K. Kato, J. Mol. Catal. B: Enzym., 2004, 30, 189–194 CrossRef CAS PubMed.
- H. Zhao, J. Chem. Technol. Biotechnol., 2010, 85, 891–907 CrossRef CAS.
- G. G. Dodson, D. P. Lane and C. S. Verma, EMBO Rep., 2008, 9, 144–150 CrossRef CAS PubMed.
- M. Karplus and J. A. McCammon, Nat. Struct. Mol. Biol., 2002, 9, 646–652 CAS.
- M. Norin, F. Haeffner, K. Hult and O. Edholm, Biophys. J., 1994, 67, 548–559 CrossRef CAS.
- H. A. Scheraga, M. Khalili and A. Liwo, Annu. Rev. Phys. Chem., 2007, 58, 57–83 CrossRef CAS PubMed.
- B. A. Tejo, A. B. Salleh and J. Pleiss, J. Mol. Model., 2004, 10, 358–366 CrossRef CAS PubMed.
- P. Trodler and J. Pleiss, BMC Struct. Biol., 2008, 8, 9 CrossRef PubMed.
- M. Klahn, G. S. Lim, A. Seduraman and P. Wu, Phys. Chem. Chem. Phys., 2011, 13, 1649–1662 RSC.
- N. M. Micaelo and C. M. Soares, J. Phys. Chem. B, 2008, 112, 2566–2572 CrossRef CAS PubMed.
- M. Klähn, G. S. Lim and P. Wu, Phys. Chem. Chem. Phys., 2011, 13, 18647–18660 RSC.
- P. R. Burney and J. Pfaendtner, J. Phys. Chem. B, 2013, 117, 2662–2670 CrossRef CAS PubMed.
- M. A. M. Latif, B. A. Tejo, R. Abedikargiban, M. B. Abdul Rahman and N. M. Micaêlo, J. Biomol. Struct. Dyn., 2013, 32(8), 1263–1273 Search PubMed.
- H. Monhemi, M. R. Housaindokht and M. S. Googheri, J. Supercrit. Fluids, 2012, 69(6), 1–7 CrossRef CAS PubMed.
- B. Mostofian, J. Smith and X. Cheng, Interdisciplinary Sciences: Computational Life Sciences, 2011, 3, 308–320 CrossRef CAS PubMed.
- C. Li, T. Tan, H. Zhang and W. Feng, J. Biol. Chem., 2010, 285, 28434–28441 CrossRef CAS PubMed.
- S. Raza, L. Fransson and K. Hult, Protein Sci., 2001, 10, 329–338 CrossRef CAS PubMed.
- V. W. Jaeger and J. Pfaendtner, ACS Chem. Biol., 2013, 8(6), 1179–1186 CrossRef CAS PubMed.
- A. Zaks and A. M. Klibanov, J. Biol. Chem., 1988, 263, 8017–8021 CAS.
- N. M. Micaêlo and C. M. Soares, FEBS J., 2007, 274, 2424–2436 CrossRef PubMed.
- N. M. Micaelo, V. H. Teixeira, A. M. Baptista and C. M. Soares, Biophys. J., 2005, 89, 999–1008 CrossRef CAS PubMed.
- J. Uppenberg, M. T. Hansen, S. Patkar and T. A. Jones, Structure, 1994, 2, 293–308 CrossRef CAS.
- P. Grochulski, Y. Li, J. D. Schrag, F. Bouthillier, P. Smith, D. Harrison, B. Rubin and M. Cygler, J. Biol. Chem., 1993, 268, 12843–12847 CAS.
- W. L. Jorgensen, D. S. Maxwell and J. Tirado-Rives, J. Am. Chem. Soc., 1996, 118, 11225–11236 CrossRef CAS.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS PubMed.
- J. N. Canongia Lopes and A. A. H. Pádua, J. Phys. Chem. B, 2006, 110, 19586–19592 CrossRef CAS PubMed.
- J. N. Canongia Lopes and A. A. H. Pádua, J. Phys. Chem. B, 2004, 108, 16893–16898 CrossRef.
- J. N. Canongia Lopes, A. A. H. Padua and K. Shimizu, J. Phys. Chem. B, 2008, 112, 5039–5046 CrossRef CAS PubMed.
- L. Martinez, R. Andrade, E. G. Birgin and J. M. Martinez, J. Comput. Chem., 2009, 30, 2157–2164 CrossRef CAS PubMed.
- A. Zaks and A. M. Klibanov, Proc. Natl. Acad. Sci. U. S. A., 1985, 82, 3192–3196 CrossRef CAS.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS PubMed.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS PubMed.
- Z. Gu and J. F. Brennecke, J. Chem. Eng. Data, 2002, 47, 339–345 CrossRef CAS.
- J. A. Laszlo and D. L. Compton, Biotechnol. Bioeng., 2001, 75, 181–186 CrossRef CAS PubMed.
- Z. Yang, Y.-J. Yue, W.-C. Huang, X.-M. Zhuang, Z.-T. Chen and M. Xing, J. Biol. Chem., 2009, 145, 355–364 CAS.
- L. Yang, J. S. Dordick and S. Garde, Biophys. J., 2004, 87, 812–821 CrossRef CAS PubMed.
- S. H. Schőfer, N. Kaftzik, P. Wasserscheid and U. Kragl, Chem. Commun., 2001, 425–426 RSC.
- J. L. Kaar, A. M. Jesionowski, J. A. Berberich, R. Moulton and A. J. Russell, J. Am. Chem. Soc., 2003, 125, 4125–4131 CrossRef CAS PubMed.
- M. T. Ru, J. S. Dordick, J. A. Reimer and D. S. Clark, Biotechnol. Bioeng., 1999, 63, 233–241 CrossRef CAS.
- M. Sandoval, A. Cortes, C. Civera, J. Trevino, E. Ferreras, M. Vaultier, J. Berenguer, P. Lozano and M. J. Hernaiz, RSC Adv., 2012, 2, 6306–6314 RSC.
- T. Zhou, L. Chen, Y. Ye, L. Chen, Z. Qi, H. Freund and K. Sundmacher, Ind. Eng. Chem. Res., 2012, 51, 6256–6264 CrossRef CAS.
- C. D. Tran, S. H. De Paoli Lacerda and D. Oliveira, Appl. Spectrosc., 2003, 57, 152–157 CrossRef CAS PubMed.
- A. M. Klibanov, Trends Biotechnol., 1997, 15, 97–101 CrossRef CAS.
- J. G. Huddleston, A. E. Visser, W. M. Reichert, H. D. Willauer, G. A. Broker and R. D. Rogers, Green Chem., 2001, 3, 156–164 RSC.
- J. D. Holbrey, W. M. Reichert and R. D. Rogers, Dalton Trans., 2004, 2267–2271 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra07460b |
| This journal is © The Royal Society of Chemistry 2014 |