Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
From in vitro to in cellulo: structure–activity relationship of (2-nitrophenyl)methanol derivatives as inhibitors of PqsD in Pseudomonas aeruginosa†
Michael P.
Storz
a,
Giuseppe
Allegretta
a,
Benjamin
Kirsch
a,
Martin
Empting
*a and
Rolf W.
Hartmann
*ab
aHelmholtz-Institute for Pharmaceutical Research Saarland (HIPS), Campus C23, 66123 Saarbrücken, Germany. E-mail: rolf.hartmann@helmholtz-hzi.de; Fax: +49 681 302 70308; Tel: +49 681 302 70300
bPharmaceutical and Medicinal Chemistry, Saarland University, Campus C23, 66123 Saarbrücken, Germany
First published on 29th May 2014
Abstract
Recent studies have shown that compounds based on a (2-nitrophenyl)methanol scaffold are promising inhibitors of PqsD, a key enzyme of signal molecule biosynthesis in the cell-to-cell communication of Pseudomonas aeruginosa. The most promising molecule displayed anti-biofilm activity and a tight-binding mode of action. Herein, we report on the convenient synthesis and biochemical evaluation of a comprehensive series of (2-nitrophenyl)methanol derivatives. The in vitro potency of these inhibitors against recombinant PqsD as well as the effect of selected compounds on the production of the signal molecules HHQ and PQS in P. aeruginosa were examined. The gathered data allowed the establishment of a structure–activity relationship, which was used to design fluorescent inhibitors, and finally, led to the discovery of (2-nitrophenyl)methanol derivatives with improved in cellulo efficacy providing new perspectives towards the application of PqsD inhibitors as anti-infectives.
Introduction
Until recently, bacterial communities were seen as nothing more than an accumulation of autonomous single-celled organisms. But today, we are aware that bacteria use cell-to-cell communication systems like quorum sensing (QS) to behave collectively rather than as individuals.1 Small diffusible molecules are produced by single bacterial cells that can be released into the environment and detected by surrounding bacteria. Upon proliferation, the extracellular signal molecule concentrations increase along with cell density. Once a certain threshold is reached, receptors are activated by these autoinducers resulting in population-wide changes in gene expression. This concerted switch from low- to high-cell-density mode allows single bacterial cells to limit their group-beneficial efforts to those cell densities which guarantee an effective group outcome. Even if QS may not be directly essential for the survival of a singular bacterial cell, it is very important for bacteria–host interactions and pathogenesis upon bacterial infections in general. In this regard, QS regulates a variety of virulence factors, which contribute to breaking the first line defences and damaging surrounding tissues leading to dissemination, systemic inflammatory-response syndrome, multiple organ failure, and, finally, death of the host.2 Furthermore, QS contributes to the collective coordination of biofilm formation, a key reason for bacterial resistance against conventional antibiotics in clinical use.3 Thus, the importance of these regulatory systems could be exploited for the design of novel anti-infectives.Several groups have successfully targeted QS, which is discussed as an alternative to the traditional treatment using bactericidal or bacteriostatic agents (for reviews see ref. 4 and 5). Novel anti-virulence compounds ideally decrease pathogenicity without affecting bacterial survival or growth, whereas it is believed that no or less selection pressure is posed on the bacteria. Hence, a reduced rate of newly occurring resistances, which gradually render existing antibiotics ineffective, is expected.6
Among bacteria, very different communication systems based on distinct autoinducers are utilized. Gram-positive bacteria primarily use modified oligopeptides, whereas N-acyl homoserine lactones are a major class of signal molecules in Gram-negative bacteria.1 The opportunistic pathogen P. aeruginosa additionally utilizes a characteristic pqs system, which is based on the quinolone PQS (Pseudomonas Quinolone Signal) and its biosynthetic precursor HHQ (2-heptyl-4-quinolone) (Fig. 1).7
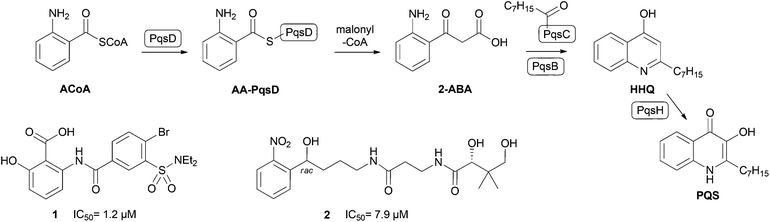 | ||
| Fig. 1 Role of PqsD in HHQ and PQS biosynthesis (top). Structures of previously reported PqsD inhibitors 1 and 2. | ||
Both are able to activate the transcriptional regulator PqsR leading to the production of various virulence factors like pyocyanin and hydrogen cyanide (HCN).8 We have shown, that PqsR antagonists efficiently decrease pyocyanin production and pathogenicity of P. aeruginosa PA14.9,10 Furthermore, HHQ and PQS contribute to the formation of biofilms.3
Biosynthesis of HHQ and PQS is accomplished by proteins encoded by the pqsABCD operon. Thereby, experiments using transposon knockout mutants identified PqsD as key enzyme in the cellular signal molecule production route.11,12 Recently, Dulcey et al. reported that cytoplasmic PqsD catalyses the condensation of anthraniloyl-CoA (ACoA) and malonyl-CoA to 2-aminobenzoylacetate (2-ABA, Fig. 1).13 The resulting reactive intermediate is then processed to HHQ by PqsC using octanoic acid as substrate. This second reaction step is supported by PqsB by an unknown mechanism. Interestingly, PqsD alone is also capable of generating HHQ in vitro directly from ACoA using β-ketodecanoic acid as secondary substrate.14 This enzymatic reaction has been routinely exploited by us to evaluate PqsD inhibitors.14–18 Inhibition of PqsD is an attractive strategy to interfere with QS-controlled infection mechanisms, since it is essential for cellular HHQ/PQS formation. A pqsD transposon mutant strain of P. aeruginosa PAO1, which is deficient in PQS formation, shows decreased levels of pyocyanin and HCN as well as reduced lethality in nematodes.11 Furthermore, putative inhibitors of PqsA, an enzyme involved in earlier stages of HHQ biosynthesis, block the cellular production of the corresponding signal molecules, prevent systemic dissemination, and attenuate mortality in infected mice.19
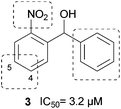
Derived from compounds active against FabH, a structurally and functionally related enzyme, we have identified and optimized the first PqsD inhibitors demonstrating IC50 values in the single-digit micromolar range (Fig. 1, 1).14,15 Unfortunately, these compounds had no pronounced effect on the extracellular signal molecule levels in cell-based assays using P. aeruginosa PA14 (unpublished data). Recently, in a ligand-based approach we have identified compound 2 as a novel inhibitor of PqsD (Fig. 1).16 Ligand efficiency-guided optimisation led to compound 3 (Fig. 2), which was used for an initial examination of the effects on PA14 cells mediated by PqsD inhibition.16 Indeed, this compound was capable of reducing the HHQ and PQS levels. Furthermore, biofilm formation was significantly inhibited and no antibiotic effects were observed.
Binding studies of 3 revealed apparent irreversibility and that binding occurs near the active site residues.17 Both enantiomers showed similar affinity but contrary thermodynamic profiles. Based on site-directed mutagenesis, isothermal titration calorimetry (ITC) analysis, and molecular docking, explicit binding modes were proposed. In these predicted enzyme–inhibitor complexes both enantiomers reside in nearly identical positions with the main difference being the orientation of the hydroxyl group at the stereogenic center.17
Herein, we present a target-oriented (in vitro) structure–activity relationship and optimization of this compound class based on the (2-nitrophenyl)methanol scaffold by systematic structure variation (Fig. 2) investigating also the time-dependent onset of inhibition. Previously, we reported, that a tetrahedral geometry including an acceptor function is favoured for the linker between both phenyl rings.16 However, the intrinsic nitrophenyl moiety bears an increased risk of toxic, mutagenic and carcinogenic side effects.20 Thus, we evaluated the mutagenicity in Ames Salmonella assays and investigated suitable chemical replacements. Additionally, the influences of substituents with opposed electronic and hydrophilic properties in 4- and 5-position of the nitrophenyl moiety were studied. Furthermore, a variety of aliphatic and aromatic residues instead of the second phenyl ring were examined.
The gathered information enabled us to design fluorescent inhibitors, which may be useful tools to investigate enzyme inhibitor interactions and to visualize the target in cells.21,22
Finally, selected compounds were examined regarding their potency to inhibit signal molecule production in P. aeruginosa PA14 cells. Thereby, we additionally applied a novel strategy to reduce costs and time by the usage of a pqsH-deficient mutant which has been selected from a transposon mutant library.23 This procedure allows to evaluate the potency of a compound solely by quantification of HHQ instead of two signal molecules. In this way we identified compounds with increased in cellulo activity while a low molecular weight is retained (<250 Da). These optimized fragment-like molecules provide the potential for further improvements by a fragment growing approach. Additionally, an attempt to correlate in vitro data with the effects observed in the cellular assays is made.
Results and discussion
Chemistry
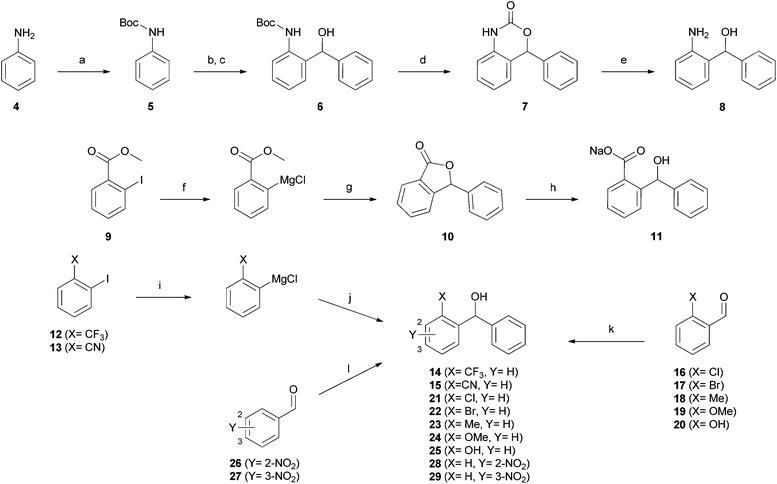
In order to find alternatives to the nitro group, a variety of molecules with different chemical functionalities were synthesized (Scheme 1). Assembly of (2-aminophenyl)(phenyl)methanol 8 started with the formation of Boc-protected aniline 5. Ortho-lithiation by tert-butyllithium and subsequent reaction with benzaldehyde yielded the alcohol 6. The desired product 8 was obtained by a two-step deprotection using trifluoroacetic acid and basic hydrolysis. This route also provided access to the cyclic carbamate 7.The carboxylate 11 was prepared by iso-propylmagnesium chloride-mediated iodine–magnesium exchange on methyl 2-iodobenzoate 9 and subsequent addition to benzaldehyde. This reaction is followed by spontaneous cyclisation yielding lactone 10, which was hydrolyzed under basic conditions to yield the desired carboxylate. An analogous method employing iodine–magnesium exchange was used for synthesis of the trifluoromethyl and nitril derivatives 14 and 15. In the case of compounds 21–25 and 28–29, corresponding aldehyde precursors were commercially available. Hence, the desired products were prepared by direct addition of phenylmagnesium chloride.
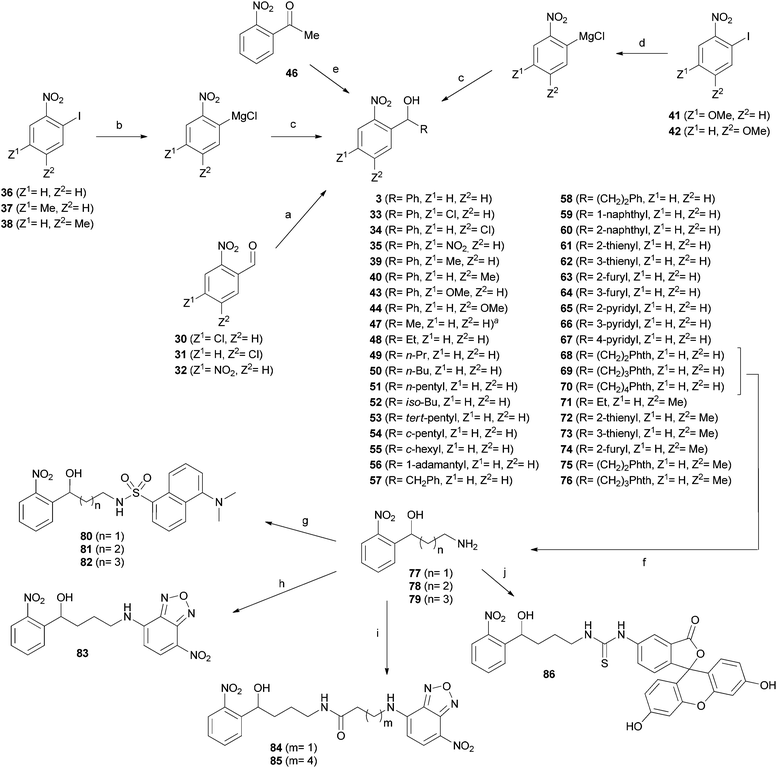
The synthesis of (2-nitrophenyl)methanol derivatives 3, 33–35, 39, 40, 43–44, and 47–86 followed the general pathways outlined in Scheme 2. For all compounds, in which Z1 or Z2 were exclusively substituted by hydrogen or methyl, phenylmagnesium chloride was added to 36–38 to accomplish iodine–magnesium exchange in ortho position to NO2 as described by Knochel and coworkers.24 The generated Grignard reagents were reacted with the appropriate aldehydes to form the desired products.
For synthesis of the methoxy derivatives 43 (Z1 = OMe) and 44 (Z2 = OMe) from 41 and 42 we utilized 4-methoxyphenylmagnesium bromide as novel reagent to accomplish iodine–magnesium exchange in ortho-iodo-nitrobenzenes. Application of this method to a broader range of substrates will be discussed elsewhere.
Fluorescent derivatives were prepared by cleavage of the phthalimide moiety of 68–70via the Ing-Manske procedure. The released amines 77–79 served as a starting point for the introduction of fluorophores. Coupling these amines with dansyl chloride yielded derivatives 80–82. Direct attachment of NBD to the amine 78 using NBD-chloride afforded 83, whereas 84 and 85 were synthesized by coupling with the NBD containing carboxylic acids. The fluorescein derivative 86 was formed upon reaction with fluorescein iso-thiocyanate.
Essentiality of the nitro-group and Ames test
After synthesis, compounds were evaluated regarding their inhibitory activity against heterologously expressed and purified PqsD using ACoA and β-ketodecanoic acid as substrates.14 Until recently, β-ketodecanoic acid instead of malonyl-CoA has been considered as the second substrate in HHQ synthesis, since it has been shown, that addition of β-ketodecanoic acid to the anthraniloyl-PqsD complex leads to HHQ formation in vitro.14 We have clearly shown that (2-nitrophenyl)methanol derivatives interfere with the formation of the anthraniloyl-PqsD complex itself, which allows further usage of β-ketodecanoic acid independently of its function in the bacterial cells.17First, a variety of substituents replacing the nitro group in ortho position were tested. An amino group (8) in analogy to ACoA, which served as template for the inhibitor design,16 led to an inactive compound. This was also true for substituents with electron-withdrawing properties similar to the nitro group, as trifluoromethyl (14), nitril (15) and halogens (21, 22). Furthermore, no activity was observed for molecules bearing potential hydrogen bond acceptors like 7, 10, the carboxylate 11, 24 and 25. Since the nitro group seems to be essential for activity, we shifted the position in meta or para position (28, 29), but inhibitory potency was again completely abolished. An initial toxicity study provided promising results, since no toxic effect against human THP-1 macrophages was observed at 250 μM of compound 3.16 To assess the mutagenic risk of the compound class, Ames Salmonella assays were performed. Compound 3 was tested on Salmonella typhimurium derived strains TA100, TA1535 and TA102 with and without metabolic activation by liver homogenate (S9 mix). No biologically relevant increase in the number of revertant colonies was observed at dose levels up to 5000 μg per plate. Thus, the nitro group was retained and we focused our efforts on the improvement of inhibitory activity by introduction of additional substituents into the nitrophenyl moiety.
In vitro SAR
Recently, we have reported extensive studies on the mode of action of the (2-nitrophenyl)methanol scaffold.17 In this regard, we demonstrated a time-dependent onset of inhibition based on a slow non-covalent (reversible) interaction. As we have observed that inhibition onset levels out after 20 min of preincubation for compound 3,17 we consider a period of 30 min appropriate for the rapid evaluation of the set of novel compounds (n ∼ 50) described herein. Additionally, we measured the inhibitory activity using only 10 min of preincubation to gain qualitative insight into the effect of inhibitor modifications on binding behaviour.This examination is relevant, as it has been reported that time-dependency of enzyme–inhibitor interactions can have significant impact in the efficacy of compounds in the cellular system.25
First, we re-evaluated our starting compound 3 applying an optimized protocol for the prolonged pre-treatment period of enzyme with inhibitor (Table 1). As described earlier,17 an improvement of potency was observed rendering this compound now a sub-micromolar PqsD inhibitor.
| Compounds | Z1 | Z2 | R | IC50 [μM] (10 min)a,b | IC50 [μM] (30 min)a,c |
|---|---|---|---|---|---|
| a P. aeruginosa PqsD (recombinantly expressed in Escherichia coli), anthraniloyl-CoA (5 μM), and β-ketodecanoic acid (70 μM). b IC50 values were determined using a 10 min preincubation period of inhibitor and enzyme followed by a 40 min reaction time. c IC50 values were determined using a 30 min preincubation period of inhibitor and enzyme followed by a 40 min reaction time. d For the structure of the fluorescent derivatives 80–86 see Scheme 2. | |||||
| 3 | H | H | Ph | 3.2 ± 0.1 | 0.5 ± 0.1 |
| 33 | Cl | H | Ph | 13.4 ± 1.4 | 11.2 ± 0.9 |
| 34 | H | Cl | Ph | 15.0 ± 0.6 | 1.6 ± 0.1 |
| 35 | NO2 | H | Ph | 15.4 ± 2.0 | 5.8 ± 1.0 |
| 39 | Me | H | Ph | 3.7 ± 0.5 | 1.6 ± 0.1 |
| 40 | H | Me | Ph | 1.9 ± 0.4 | 0.6 ± 0.1 |
| 43 | OMe | H | Ph | 2.2 ± 0.5 | 1.6 ± 0.3 |
| 44 | H | OMe | Ph | 3.0 ± 0.4 | 0.5 ± 0.1 |
| 45 | H | H | H | 1.6 ± 0.5 | 0.7 ± 0.2 |
| 47 | H | H | Me | 1.3 ± 0.3 | 0.8 ± 0.1 |
| 48 | H | H | Et | 1.1 ± 0.2 | 0.8 ± 0.1 |
| 49 | H | H | n-Pr | 2.8 ± 0.4 | 1.0 ± 0.4 |
| 50 | H | H | n-Bu | 5.2 ± 0.8 | 2.9 ± 0.1 |
| 51 | H | H | n-Pentyl | 4.9 ± 0.9 | 1.0 ± 0.4 |
| 52 | H | H | iso-Bu | 7.9 ± 1.0 | 1.2 ± 0.3 |
| 53 | H | H | tert-Pentyl | 15.9 ± 1.1 | 6.7 ± 0.1 |
| 54 | H | H | c-Pentyl | 4.9 ± 1.0 | 1.4 ± 0.2 |
| 55 | H | H | c-Hexyl | 10.1 ± 1.4 | 4.5 ± 0.2 |
| 56 | H | H | 1-Adamantyl | 11.6 ± 2.2 | 2.7 ± 0.1 |
| 57 | H | H | CH2Ph | 5.4 ± 0.6 | 0.8 ± 0.1 |
| 58 | H | H | CH2CH2Ph | 4.6 ± 1.0 | 0.9 ± 0.1 |
| 59 | H | H | 1-Naphthyl | 10.8 ± 2.5 | 5.8 ± 0.2 |
| 60 | H | H | 2-Naphthyl | 13.1 ± 1.9 | 2.4 ± 0.5 |
| 61 | H | H | 2-Thienyl | 14.3 ± 1.9 | 1.5 ± 0.2 |
| 62 | H | H | 3-Thienyl | 5.9 ± 0.9 | 6.4 ± 1.9 |
| 63 | H | H | 2-Furyl | 1.8 ± 0.4 | 0.9 ± 0.1 |
| 64 | H | H | 3-Furyl | 28% @ 50 μM | 13.1 ± 2.8 |
| 65 | H | H | 2-Pyridyl | 6.7 ± 1.2 | 1.2 ± 0.1 |
| 66 | H | H | 3-Pyridyl | 11.7 ± 2.1 | 1.7 ± 0.1 |
| 67 | H | H | 4-Pyridyl | 6.8 ± 1.4 | 2.2 ± 0.1 |
| 68 | H | H | (CH2)2Phth | 1.2 ± 0.1 | 0.3 ± 0.1 |
| 69 | H | H | (CH2)3Phth | 1.9 ± 0.3 | 0.7 ± 0.2 |
| 70 | H | H | (CH2)4Phth | 1.7 ± 0.5 | 1.6 ± 0.4 |
| 71 | H | Me | Et | 0.7 ± 0.3 | 0.6 ± 0.1 |
| 72 | H | Me | 2-Thienyl | 6.2 ± 1.8 | 2.7 ± 0.5 |
| 73 | H | Me | 3-Thienyl | 3.3 ± 0.5 | 1.6 ± 0.3 |
| 74 | H | Me | 2-Furyl | 0.9 ± 0.1 | 1.1 ± 0.1 |
| 75 | H | Me | (CH2)2Phth | 0.9 ± 0.1 | 0.7 ± 0.1 |
| 76 | H | Me | (CH2)3Phth | 0.7 ± 0.1 | 0.7 ± 0.2 |
| 77 | H | H | (CH2)2NH2 | 34.7 ± 4.6 | 40.0 ± 3.7 |
| 78 | H | H | (CH2)3NH2 | 44% @ 50 μM | 27.8 ± 1.7 |
| 79 | H | H | (CH2)4NH2 | 8.2 ± 2.1 | 22.4 ± 4.3 |
| 80 | 3.5 ± 0.1 | 5.8 ± 0.2 | |||
| 81 | 3.0 ± 1.3 | 4.0 ± 0.7 | |||
| 82 | 3.2 ± 0.4 | 1.4 ± 0.3 | |||
| 83 | 4.3 ± 0.5 | 3.4 ± 0.2 | |||
| 84 | 46.5 ± 2.1 | 12.5 ± 4.2 | |||
| 85 | 13.1 ± 1.5 | 10.0 ± 0.7 | |||
| 86 | 1.3 ± 0.7 | 1.5 ± 0.3 | |||
In a subsequent step, we investigated the effect of different substituents within the nitrophenyl moiety. Compounds bearing electron-withdrawing substituents (EWG) as chlorine (33, 34) or nitro (35) showed diminished inhibitory activity (Table 1). However, concerning target affinity (IC50 at 30 min of preincubation) the para substitution pattern (Z2 in Table 1) seems to be more favourable. In contrast, introduction of the electron-donating (EDG) methyl (39, 40) or methoxy (43, 44) groups led to potent compounds with IC50 values in the range of the unsubstituted 3. Again, a preference for the introduction of substituents at Z2 was observed (40, 44). These observations are in accordance with our proposed binding mode reported earlier,17 as the para position to the nitro group provides more space to accommodate additional substituents. An explanation for the general detrimental effect of EWGs on affinity could be that the nitro group functions as hydrogen bond acceptor (as in our proposed binding model). This ability might possibly be diminished by electron-withdrawing substituents. Unfortunately, none of the modifications installed in this part of the scaffold led to an improvement of inhibitor potency. However, comparing the determined IC50 for 10 min and 30 min, it seems that the Z2 position provides the opportunity to modulate the binding behaviour. Through the choice of either methyl (EDG) or chloro (EWG) substituents, the onset (see IC50 at 10 min) can be either slightly accelerated (3vs.40) or slowed down (3vs.34).
In light of the results gathered so far, we kept the unsubstituted nitrophenyl moiety constant and turned our attention to the second residue R of the methanol moiety. Hydrogen or linear alkyl chains of different length were introduced (45, 47–51). Thereby, shorter residues up to ethyl (45, 47, and 48) were favoured over longer variants (49–51). A plausible explanation is the increasing entropic penalty caused by the limitation of rotational freedom upon formation of the inhibitor–enyzme complex.
Except iso-butyl-bearing compound 52, branched and cyclic isomers 53–56 generally inhibited PqsD less efficiently than their linear congeners. This might be due to the narrow entrance channel, which hampers the binding of the bulky residues.
Short alkyl linkers were inserted between the tetrahedral carbon and the phenyl group, but neither compound 57 nor 58 showed improved IC50 values compared to 3. Thus, we concluded that direct attachment to the methanol moiety brings the aromatic residue in an optimal position and fused a second benzene ring. But the resulting 1-naphthyl and 2-naphthyl isomers 59 and 60 were less potent PqsD inhibitors.
Hence, monocyclic heteroaromatic residues were introduced. For all the thiophene and pyridine derivatives 61, 62, and 65–67 moderate activity without further improvement was observed. The furane derivatives showed conspicuous behaviour, since the differences in activity between the oxygen in 2- or 3-position were tremendous. While the 3-furyl derivative 64 was almost inactive, the 2-furyl isomer 63 shows improved PqsD inhibition. The synthetic route towards fluorescent (2-nitrophenyl)methanol derivatives provided additional inhibitors of PqsD with non-fluorescent residues R as intermediates. The amines 77–79 may be considered as direct derivatives of the alkyl compounds 49–51, whereas the terminal methyl was substituted by an amino group, which is expected to be protonated under assay conditions. In contrast to the alkyl compounds, the amines 77–79 showed low activity. This result was expected, as the entrance of the substrate tunnel is decorated with arginine side chains providing a repulsive positive surface polarization.
The phthalimides 68–70, on the other hand, also differing in the length of the alkyl linker, showed potent PqsD inhibition. Compound 68 even demonstrated the lowest IC50 value within the investigated set of compounds of around 300 nM.
So far, novel interesting derivatives with retained potency and reduced molecular weight (45, 47, 48, and 63) or even improved target affinity (68) have been identified. Moreover, some compounds showed a pronounced difference between the IC50 values measured via the 10 min and 30 min protocol (34, 52, 61, and 66) indicating a slow onset of inhibition. As described above, introduction of a methyl group in para position to the nitro group resulted in a reduction of time-dependency while retaining activity for compound 40. These results encouraged us to synthesize additional selected derivatives possessing this methyl group (71, 72). Indeed, all of these methyl-containing compounds showed a fast onset of inhibition (Table 1) while being potent inhibitors of PqsD in the single-digit micromolar to submicromolar range. However, no further improvement in target affinity has been gained compared to the most potent compound 68. Nevertheless, together with the unmethylated congeners (48, 61–63, 68, and 69) interesting pairs of PqsD inhibitors for further evaluation in cellulo have been yielded.
Apparently, various substituents of R are tolerated by PqsD, which encouraged us to introduce fluorescent groups in this position. Since promising inhibitory activity was observed for compound 2, we substituted the pantothenate moiety by dansyl (5-(dimethylamino)-naphthalene-1-sulfonyl), NBD (7-nitrobenz-2-oxa-1,3-diazol-4-yl) and fluorescein fluorophores. The flexible linker of 2 was conserved to provide sufficient degrees of conformational freedom to adopt to the sterical requirements of the substrate tunnel. In the case of the dansyl derivatives 80–82, the chain length of the alkyl linker was only slightly varied with the intention to position the hydrophobic fluorophore within the channel. For the more hydrophilic NBD derivatives 83–85 additional acyl linkers were introduced as well, thereby shifting the fluorophore towards the protein surface. Fortunately, an acceptable in vitro activity was observed for the dansyl derivatives 80–82, and compounds with NBD (83) or fluorescein (86) directly attached to the amine 78. However, the NBD fluorophores 84 and 85 containing an additional acyl-linker were less potent. Unfavourable interactions and/or entropic penalties might be possible reasons.
Inhibition of signal molecule production in a P. aeruginosa pqsH mutant
In an attempt to elucidate the physicochemical and/or functional requirements for high in cellulo efficacy, we tested selected compounds regarding their ability to reduce the signal molecule levels in P. aeruginosa PA14 (Table 2).| Cmpd | MWa | Log Pa,b | LEa,c | IC50 (10 min)/IC50 (30 min)a | % HHQ inhibitiond |
|---|---|---|---|---|---|
| a Molecular weight (MW), calculated partition coefficient (log P), ligand efficiency index (LE), and the ratio of IC50 values measured using different preincubation periods. b Calculated by ACD/Labs 2012 using the ACD/Log P Classic algorithm. c Values calculated as LE = 1.4 (−log IC50)/N using the IC50 value for the prolonged incubation time (30 min) and N meaning number of non-hydrogen atoms. d Planctonic P. aeruginosa PA14 pqsH mutant. Inhibitor concentration 250 μM as not indicated otherwise. Percentage of inhibition was normalized regarding OD600. n.i. no significant inhibition (<10%). e n.d. means “not determined”. | |||||
| 3 | 229.23 | 2.47 | 0.52 | 6.4 | 43 ± 6 |
| 33 | 263.68 | 3.16 | 0.39 | 1.2 | n.i. |
| 34 | 263.68 | 3.12 | 0.45 | 9.4 | n.i. |
| 35 | 274.23 | 2.14 | 0.37 | 2.7 | 10 ± 4 |
| 39 | 243.26 | 2.93 | 0.45 | 2.3 | 20 ± 3 |
| 40 | 243.26 | 2.93 | 0.48 | 3.2 | 19 ± 7 |
| 43 | 259.26 | 2.69 | 0.43 | 1.4 | n.i. |
| 44 | 259.26 | 2.55 | 0.46 | 6.0 | n.i. |
| 45 | 153.14 | 0.76 | 0.78 | 2.3 | 13 ± 1 |
| 47 | 167.16 | 1.11 | 0.71 | 1.6 | 26 ± 6 |
| 48 | 181.19 | 1.64 | 0.66 | 1.4 | 61 ± 2 |
| 49 | 195.22 | 2.17 | 0.60 | 2.8 | 22 ± 7 |
| 50 | 209.24 | 2.71 | 0.52 | 1.7 | n.i. |
| 51 | 223.27 | 3.24 | 0.53 | 4.9 | n.i. |
| 61 | 235.26 | 2.15 | 0.51 | 8.7 | 64 ± 6 |
| 62 | 235.26 | 2.15 | 0.45 | 0.8 | 74 ± 6 |
| 63 | 219.19 | 1.63 | 0.53 | 2.0 | 51 ± 15 |
| 64 | 219.19 | 1.63 | 0.43 | n.d.e | 73 ± 2 |
| 68 | 326.30 | 2.61 | 0.38 | 4.0 | n.i. @ 125 μM |
| 69 | 340.33 | 2.84 | 0.34 | 2.7 | n.i. @ 125 μM |
| 70 | 354.36 | 2.98 | 0.31 | 1.1 | n.i. @ 125 μM |
| 71 | 195.22 | 2.10 | 0.62 | 1.2 | n.i. |
| 72 | 249.24 | 2.61 | 0.46 | 2.3 | 24 ± 0.2 |
| 73 | 249.24 | 2.61 | 0.48 | 2.1 | 23 ± 7 |
| 74 | 233.22 | 2.09 | 0.49 | 0.8 | 20 ± 2 |
| 75 | 340.33 | 3.07 | 0.34 | 1.3 | n.i. @ 125 μM |
| 76 | 354.36 | 3.30 | 0.33 | 1.0 | n.i. @ 100 μM |
| 80 | 429.49 | 3.71 | 0.24 | 0.6 | n.i. @ 75 μM |
| 81 | 443.52 | 3.94 | 0.24 | 0.8 | n.i. @ 75 μM |
| 82 | 457.54 | 4.08 | 0.26 | 2.3 | n.i. @ 25 μM |
| 83 | 373.32 | 1.10 | 0.28 | 1.3 | n.i. @ 150 μM |
| 84 | 444.40 | 2.15 | 0.21 | 3.7 | 17 ± 3 |
| 85 | 486.48 | 2.65 | 0.20 | 1.3 | 27 ± 1 |
| 86 | 599.61 | 2.94 | 0.19 | 0.9 | n.i. |
In the wild-type strain, HHQ is converted into PQS by PqsH and the expression of pqsH depends on the growth period.26 Thus, we quantified HHQ in a pqsH mutant, which is not able to perform this oxidation, to increase simplicity and reproducibility of this cell-based assay. To ensure the validity of this novel methodology, we compared the results gathered using the pqsH mutant with additionally determined PA14 wild-type data for three selected compounds and observed a good correlation between both data sets (see ESI†).
If soluble, all compounds were tested at 250 μM. The in cellulo results are summarized in Table 2 together with relevant parameters like molecular weight (MW), calculated partition coefficient (log P), ligand efficiency index (LE), and the ratio of IC50 values measured using different preincubation periods.
Inhibitor 3, which served as a starting point, decreased HHQ production by 43%. Introduction of methyl groups into the nitro-phenyl moiety (39 and 40) reduced inhibitory potency, whereas methoxy substituents (43 and 44) led to inactivity. These results are disappointing, since two of the compounds showed submicromolar in vitro activity. A similar result was obtained for molecules bearing a substituent in the nitrophenyl moiety: the chloro compounds 33 and 34 as well as the dinitrophenyl-derivative 35 showed no or only low activity in the cells. Furthermore, no effects on HHQ production despite of most promising IC50 values were observed for the phthalimides 68–70, 75, and 76, which were tested at 100–125 μM due to limited solubility.
All congeners of the homologous series 45–51 containing unbranched alkyl residues showed good activity in the enzymatic assay, whereas the in vitro activity slightly decreased for the larger alkanes. In the cellular test system, the largest residues (50 and 51) led to inactivity, while a maximum HHQ inhibition was observed for ethyl (48). This compound showed an improved cellular activity compared to the starting point 3. Interestingly, the variant with methyl in para position to the nitro group (71) was inactive, although it possesses a slightly improved IC50 value.
Surprisingly, all four compounds containing heteroaromatic pentacycles 61–64 potently reduced the HHQ formation. These compounds were not among the most potent PqsD inhibitors in vitro. Moreover, 64 showed only a moderate activity in the double-digit micromolar range against recombinant PqsD. We can only speculate whether additional alternative cellular targets are involved or whether the furanyl residue is converted into a more active compound inside the cell. Multiple examples for the instability of furan are reported in the literature. However, the thiophene 62 is the most potent inhibitor of cellular HHQ formation reported so far. Again, the methyl modification in Z2 of the nitrophenyl moiety resulting in compounds 72–74 was detrimental to in cellulo efficacy.
Finally, we examined the effect of fluorophore-labelled derivatives 80–86 on P. aeruginosa to evaluate their potential for applications in cellular systems. Unfortunately, neither the dansyl- nor fluorescein-labelled inhibitors (80–82 and 86) were able to reduce HHQ formation at 250 μM. Consequently, their application is restricted to studies using lysed cells or isolated biomolecules. The best results were obtained for NBD derivatives 84 and 85, which showed at least slight inhibition. This opens up avenues towards intracellular labelling experiments, which might be conducted in the future.
Interpretation of in vitro and in cellulo data
A first conclusion which can be drawn from the detailed experimental data reported herein is that in the case of (2-nitrophenyl)methanol derivatives in vitro potency expressed either as IC50 or ligand efficiency (LE, Table 2) does not directly translate into in cellulo activity. However, this observation is not utterly surprising as Gram-negative bacteria, in general, and P. aeruginosa, in particular, are known to provide challenging barriers for the effective inhibition of intracellular targets.27–29 The orthogonal sieving ability of the two bacterial cell membranes blocking larger hydrophobic compounds (outer membrane) as well as smaller hydrophilic entities (inner membrane) from entering the cytoplasm is only one obstacle which an anti-infective agent has to overcome.27,28 Additionally, an arsenal of efflux pumps and degrading/metabolizing enzymes may hinder a drug from reaching its target.29 Hence, the physicochemical, structural, and functional features of compounds displaying intracellular activity are all the more worthwhile to investigate.In this context, time-dependent inhibition was one characteristic of the presented compound class that has first drawn our attention. Reversible target interaction (inhibition) is characterized by inhibitor binding (kon, “on-rate”) and dissociation (koff, “off-rate”). Thus, a slow binding behaviour in combination with promising (nanomolar) IC50 values would imply long drug-residence times as the “off-rate” should also be attenuated along with the inhibition onset (“on-rate”). Such an effective (long-lasting) enzyme blockade can be considered a favourable scenario in order to shut down cellular signal molecule synthesis. However, we were not able to establish a correlation between this phenomenon and improved HHQ reduction. Some compounds with either high or low ratios of IC50 values measured using 10 min or 30 min of preincubation were effective inhibitors of signal molecule production (compare for example 3 and 61 with 62 and 70). Indeed, we showed that the methyl modification at the nitrophenyl core results in compounds with only low time-dependent onset of inhibition along with a decreased or even abolished in cellulo efficacy (48vs.71 and 61vs.72). Nevertheless, the most potent inhibitor in our cell assays was 62 showing almost no time-dependency of PqsD inhibition.
A parameter which seems to have direct influence on intracellular activity, though, was molecular weight. Most compounds with MW > 300 Da were inactive inside the cells. The only exceptions within the set of tested compounds were NBD-tagged fluorophores 84 and 85 showing only a moderate reduction of HHQ production. This observed mass cut-off is much lower than the general value of 600 Da reported for Gram-negative bacteria and should not be considered a strict criteria due exceptions mentioned above.30 This finding might be explained by the described low permeability of outer membrane of P. aeruginosa compared to other Gram-negative bacteria.31
Another important physicochemical parameter for cellular availability is hydrophobicity usually expressed as the octanol–water partition coefficient log P. None of the fragment-like compounds reported herein can be considered as strongly lipophilic as none of them exceeds log P ≈ 4 while the majority possesses a value below 3. However, it has been proposed by others that even lower log P values are beneficial or actually a prerequisite for activity against Gram-negative bacteria.30 Indeed, our most active compound in cellulo possess a log P below 2.5.
Noteworthy, we have identified compounds within the set of tested (2-nitrophenyl)methanol derivatives that are interesting exceptions to the described MW/log P criteria. Compound 71, for example, has low molecular weight (195.22 Da) and log P (0.62) combined with a substantial in vitro activity (IC50 = 0.6) but no relevant in cellulo activity. We account this finding to the general detrimental effect of the methyl group in para position to the nitro group which we have also found for the other compounds bearing this substitution pattern. On the other hand, developed NDB-tagged inhibitors possess increased molecular weight and hydrophobicity, yet show moderate activity on signal molecule production. Hence, further optimization of our fragment-like molecules towards drug-like compounds via fragment growing approaches may be a rewarding endeavour.
Conclusions
More than fifty derivatives of (2-nitrophenyl)phenylmethanol 3 have been synthesized and tested for in vitro activity. In this regard, we demonstrated that 4-methoxyphenylmagnesium bromide is a suitable reagent to accomplish efficient I–Mg exchange for compound 42. Whereas the nitro group in ortho position turned out to be essential for in vitro PqsD inhibition, no mutagenicity was observed for compound 3 in an Ames test. Improved potency was achieved by the replacement of the eastern phenyl residue. This position turned out to be very tolerant for various moieties, which allowed the design of fluorescence-labelled inhibitors.For a straightforward evaluation of PqsD inhibitors in the cellular context, signal molecule production was investigated using a pqsH-deficient P. aeruginosa PA14 strain. Some of our compounds showed significantly improved inhibition of signal molecule production. An attempt to correlate in vitro data with in cellulo results has been made identifying low molecular weight and hydrophobicity as important, but not stringent, criteria for intracellular activity. Nevertheless, the presented work emphasizes the notion that optimization of intracellular activity is a challenging multi-parameter problem which requires further intense research.
Finally, novel PqsD inhibitors presented in this work possess a fragment-like size and improved efficiency in cellulo. Together with the structural insight provided by our studies regarding the mode of action, we have delivered the basis for a fragment-growing optimization process towards PqsD-targeting anti-infectives.
Experimental section
General
(2-Nitrophenyl)methanol 45 was purchased from Sigma-Aldrich and used for biological assays without further purification. Starting materials were purchased from ABCR, Acros, Sigma-Aldrich and Fluka and were used without further purification. All reactions were conducted under a nitrogen atmosphere. During workup drying was achieved by anhydrous sodium sulfate. Flash chromatography was performed using silica gel 60 (40–63 μm) and the reaction progress was determined by TLC analysis on ALUGRAM SIL G/UV254 (Macherey-Nagel). Visualization was accomplished through excitation using UV light. Purification by semi-preparative HPLC was carried out on an Agilent 1200 series HPLC system from Agilent Technologies, using an Agilent Prep-C18 column (30 × 100 mm/10 μm) as stationary phase with acetonitrile–water as eluent. The purity of compounds used in the biological assays was ≥95% as measured by LC/MS, monitored at 254 nm. The methods for LC/MS analysis and a table with analytical data (including melting points) for all tested compounds are provided in the purity section of the ESI.† All chiral alcohols were isolated as racemates. 1H and 13C NMR spectra were recorded on a Bruker DRX-500 instrument at 300 K. Chemical shifts are reported in δ values (ppm) and the hydrogenated residues of deuterated solvents were used as internal standard (acetone-d6: 2.05, 29.84. CDCl3: δ = 7.26, 77.16. MeOH-d4: δ = 3.31, 49.00. DMSO-d6: δ = 2.50, 39.52). Splitting patterns describe apparent multiplicities and are designated as s (singlet), d (doublet), dd (doublet of doublet), t (triplet), td (triplet of doublet), q (quartet), m (multiplet). All coupling constants (J) are given in hertz (Hz). Mass spectra (ESI) were measured on a Thermo Scientific Orbitrap. Mass spectra (EI) were measured on a DSQII instrument (ThermoFisher). Melting points of samples were determined in open capillaries using a SMP3 Melting Point Apparatus from Bibby Sterilin and are uncorrected. Infrared spectra were measured on a PerkinElmer Spectrum 100 FT-IR spectrometer.General method A for synthesis of 3, 39, 40, 48–76
A solution of 2-iodo-nitrobenzene 36 or a close derivative (1.0 eq.) in THF (10 ml g−1 reagent) was cooled to −40 °C and a solution of phenylmagnesium chloride (2 M in THF, 1.1 eq.) was added dropwise. After stirring for 30 min at −40 °C, the aldehyde (1.0 eq.) was added. Then, the reaction mixture was continuously stirred at −40 °C until complete conversion (checked by TLC). The reaction was quenched with a saturated solution of NH4Cl (5 ml) and diluted with water (5 ml). The aqueous phase was extracted with ethyl acetate (three times) and the combined organic layers were washed with brine, dried, and concentrated under reduced pressure. The residue was purified by column chromatography over silica gel to give the desired product.General method B for synthesis of 10, 14, and 15
A solution of the iodobenzene 9, 12 or 13 (1.00 g, 1.0 eq.) in 30 ml THF was cooled to −40 °C and a solution of iso-propylmagnesium chloride (2 M in THF, 1.1 eq.) was added dropwise. The solution was stirred for 60 min at −40 °C, benzaldehyde (1.0 eq.) was added and the reaction was completed at −40 °C (checked by TLC). The workup was carried out as described in method A.General method C for synthesis of 21–25
To a solution of phenylmagnesium chloride (2 M in THF, 1.5 eq.) in 8 ml THF at 0 °C aldehydes 16–20 (1 eq.) were slowly added and the solution was stirred at 50 °C for 30 min. The mixture was cooled to 0 °C, quenched with a saturated solution of NH4Cl (5 ml), and diluted with water (5 ml). The aqueous phase was extracted with diethyl ether (three times) and the combined organic layers were dried and concentrated under reduced pressure. The residue was purified by column chromatography over silica gel to give the desired product.General method D for synthesis of 28 and 29
To a solution of phenylmagnesium chloride (3.31 ml, 2 M in THF, 6.62 mmol) in 20 ml THF at −40 °C nitrobenzaldehyde 26 or 27 (1.00 g, 6.62 mmol) was added slowly. The reaction mixture was stirred continuously at −40 °C until complete conversion (checked by TLC). The mixture was quenched with a saturated solution of NH4Cl (5 ml) and diluted with water (5 ml). The workup was carried out as described in method A, followed by purification of the crude product by flash chromatography (petroleum ether–ethyl acetate 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
General method E for synthesis of 77–79
To a solution of the phthalimides 68–70 (1.53 mmol, 1 eq.) in ethanol (40 ml) hydrazine hydrate (9.18 mmol, 6 eq.) was added and the mixture was refluxed for 3 h. Ethanol was removed in vacuo and ethyl acetate was added. The solution was washed with water and extracted twice with EtOAc. The organic phase was dried and the solvent was evaporated to yield the desired product in sufficient purity.Expression and purification of recombinant PqsD
Expression and purification of recombinant PqsD was conducted as previously described.16 Briefly, BL21 (λDE3) E. coli transformed with expression vector pT28b(+)/pqsD were induced with IPTG overnight. After harvesting and lysis through sonication, recombinant PqsD possessing a His6-tag was isolated via immobilized metal ion affinity chromatography (IMAC) followed by gel filtration. The affinity tag was removed by thrombin cleavage and a second IMAC step.Enzyme inhibition assay using recombinant PqsD
The standard assay for determination of IC50 values was performed monitoring the enzyme activity by measuring the HHQ concentration as described recently.14 PqsD was preincubated with inhibitor for 10 min or 30 min prior to addition of the substrates ACoA and β-ketodecanoic acid. Quantification of HHQ was performed analogously, but with some modifications: The flow rate was set to 750 μl min−1 and an Accucore RP-MS column, 150 × 2.1 mm, 2.6 μm, (Thermo Scientific) was used. All test compound reactions were performed in sextuplicate. Synthesis of ACoA and β-ketodecanoic acid was performed as described in the ESI.†Cultivation of P. aeruginosa PA14 pqsH mutant
For determination of extracellular HHQ levels, cultivation was performed in the following way: cultures of P. aeruginosa PA14 pqsH transposon mutant23 (initial OD600 = 0.02) were incubated with or without inhibitor (final DMSO concentration 1%, v/v) at 37 °C, 200 rpm and a humidity of 75% for 16 h in 24-well Greiner Bio-One Cellstar plates (Frickenhausen, Germany) containing 1.5 ml medium per well. Cultures were generally grown in PPGAS medium (20 mM NH4Cl, 20 mM KCl, 120 mM Tris-HCl, 1.6 mM MgSO4, 0.5% (w/v) glucose, 1% (w/v) Bacto™ Tryptone). For each sample, cultivation and sample work-up were performed in triplicates.Determination of extracellular HHQ levels
Extracellular levels of HHQ were determined according to the method of Lépine et al. with the following modifications.32,33 An aliquot of 500 μl of bacterial cultures were supplemented with 50 μl of a 10 μM methanolic solution of the internal standard (IS) 5,6,7,8-tetradeutero-2-heptyl-4(1H)-quinolone (HHQ-d4) and extracted with 1 ml of ethyl acetate by vigorous shaking. After centrifugation, 400 μl of the organic phase were evaporated to dryness in LC glass vials. The residue was re-dissolved in methanol. UHPLC-MS/MS analysis was performed as described in detail recently.16 The following ions were monitored (mother ion [m/z], product ion [m/z], scan time [s], scan width [m/z], collision energy [V], tube lens offset [V]): HHQ: 244, 159, 0.5, 0.01, 30, 106; HHQ-d4 (IS): 248, 163, 0.1, 0.01, 32, 113. Xcalibur software was used for data acquisition and quantification with the use of a calibration curve relative to the area of the IS.Calculation of log P values
Experimental values of 67 and 79 were determined by Sirius T3 titrator from Sirius Analytical. Comparison with values calculated by ACD/Labs 2012 (Build 1996, 31. May 2012) revealed, that ACD/Log P Classic is the most appropriate algorithm. Consequently, the latter was used for the values given in Table 2.Abbreviations
| 2-ABA | 2-Aminobenzoylacetate |
| ACoA | Anthraniloyl-CoA |
| dansyl | 5-(Dimethylamino)-naphthalene-1-sulfonyl |
| EDG/EWG | Electron-donating/withdrawing group |
| HHQ | 2-Heptyl-4-quinolone |
| LE | Ligand efficiency |
| log P | Octanol–water partition coefficient |
| MW | Molecular weight |
| NBD | 7-Nitrobenz-2-oxa-1,3-diazol-4-yl |
| PQS | 2-Heptyl-3-hydroxy-4-quinolone |
| QS | Quorum sensing |
Acknowledgements
We thank Carina Scheid, Simone Amann, and Christine Maurer for performing the PqsD and cellular assays. Furthermore, we thank Prof. Dr Susanne Häußler for kindly supplying the P. aeruginosa PA14 wild-type strain and PA14 pqsH mutant and Michael Hoffmann for performing HRMS measurements.Notes and references
- S. Swift, J. A. Downie, N. A. Whitehead, A. M. Barnard, G. P. Salmond and P. Williams, Adv. Microb. Physiol., 2001, 45, 199 CrossRef CAS.
- For a review see: L. C. M. Antunes, R. B. R. Ferreira, M. M. C. Buckner and B. B. Finlay, Microbiology, 2010, 156, 2271 CrossRef CAS PubMed.
- L. Yang, M. Nilsson, M. Gjermansen, M. Givskov and T. Tolker-Nielsen, Mol. Microbiol., 2009, 74, 1380 CrossRef CAS PubMed.
- T. B. Rasmussen and M. Givskov, Microbiology, 2006, 152, 895 CrossRef CAS PubMed.
- J. C. A. Janssens, S. C. J. De Keersmaecker, D. E. De Vos and J. Vanderleyden, Curr. Med. Chem., 2008, 15, 2144 CrossRef CAS.
- For a critical discussion see: T. Defoirdt, N. Boon and P. Bossier, PLoS Pathog., 2010, 6, e1000989 Search PubMed.
- E. C. Pesci, J. B. Milbank, J. P. Pearson, S. L. McKnight, A. Kende, E. P. Greenberg and B. Iglewski, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11229 CrossRef CAS.
- J.-F. Dubern and S. P. Diggle, Mol. BioSyst., 2008, 4, 882 RSC.
- C. Lu, B. Kirsch, C. Zimmer, J. C. de Jong, C. Henn, C. K. Maurer, M. Müsken, S. Häussler, A. Steinbach and R. W. Hartmann, Chem. Biol., 2011, 19, 381 CrossRef PubMed.
- C. Lu, C. K. Maurer, B. Kirsch, A. Steinbach and R. W. Hartmann, Angew. Chem., Int. Ed., 2014, 53, 1109 CrossRef CAS PubMed.
- L. A. Gallagher, S. L. McKnight, M. S. Kuznetsowa, E. C. Pesci and C. Manoil, J. Bacteriol., 2002, 184, 6472 CrossRef CAS.
- Y.-M. Zhang, M. W. Frank, K. Zhu, A. Mayasundari and C. O. Rock, J. Biol. Chem., 2008, 283, 28788 CrossRef CAS PubMed.
- C. E. Dulcey, V. Dekimpe, D.-A. Fauvelle, S. Milot, M.-C. Groleau, N. Doucet, L. G. Rahme, F. Lépine and E. Déziel, Chem. Biol., 2013, 20, 1481 CrossRef CAS PubMed.
- D. Pistorius, A. Ullrich, S. Lucas, R. W. Hartmann, U. Kazmaier and R. Müller, ChemBioChem, 2011, 12, 850 CrossRef CAS PubMed.
- E. Weidel, J. C. de Jong, C. Brengel, M. P. Storz, A. Braunshausen, M. Negri, A. Plaza, A. Steinbach, R. Müller and R. W. Hartmann, J. Med. Chem., 2013, 56, 6146 CrossRef CAS PubMed.
- M. P. Storz, C. K. Maurer, C. Zimmer, N. Wagner, C. Brengel, J. C. de Jong, S. Lucas, M. Müsken, S. Häussler, A. Steinbach and R. W. Hartmann, J. Am. Chem. Soc., 2012, 134, 16143 CrossRef CAS PubMed.
- M. P. Storz, C. Brengel, E. Weidel, M. Hoffmann, K. Hollemeyer, A. Steinbach, R. Müller, M. Empting and R. W. Hartmann, ACS Chem. Biol., 2013, 8, 2794 CrossRef CAS PubMed.
- J. H. Sahner, C. Brengel, M. P. Storz, M. Groh, A. Plaza, R. Müller and R. W. Hartmann, J. Med. Chem., 2013, 56, 8656 CrossRef CAS PubMed.
- B. Lesic, F. Lépine, E. Déziel, J. Zhang, Q. Zhang, K. Padfield, M.-H. Castonguay, S. Milot, S. Stachel, A. A. Tzika, R. G. Tompkins and L. G. Rahme, PLoS Pathog., 2007, 3, e126 Search PubMed.
- R. Padda, C. Wang, J. Hughes, R. Kutty and G. Bennett, Environ. Toxicol. Chem., 2003, 22, 2293 CrossRef CAS.
- S. Xu, A. N. Butkevich, R. Yamada, Y. Zhou, B. Debnath, R. Duncan, E. Zandi, N. A. Petasis and N. Neamati, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16348 CrossRef CAS PubMed.
- K. A. Kozlowski, F. H. Wezeman and R. M. Schultz, Proc. Natl. Acad. Sci. U. S. A., 1984, 81, 1135 CrossRef CAS.
- N. T. Liberati, J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei and F. M. Ausubel, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 2833 CrossRef CAS PubMed.
- I. Sapountzis and P. Knochel, Angew. Chem., Int. Ed., 2002, 41, 1610 CrossRef CAS.
- R. A. Copeland, D. L. Pompliano and T. D. Meek, Nat. Rev. Drug Discovery, 2006, 5, 730 CrossRef CAS PubMed.
- E. Déziel, F. Lépine, S. Milot, J. He, M. N. Mindrinos, R. Tompkins and L. G. Rahme, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1345 CrossRef PubMed.
- L. L. Silver, Clin. Microbiol. Rev., 2011, 24, 71 CrossRef CAS PubMed.
- H. Nikaido, Antimicrob. Agents Chemother., 1989, 33, 1831 CrossRef CAS.
- K. Poole, J. Mol. Microbiol. Biotechnol., 2001, 3, 255 CAS.
- R. O'Shea and H. E. Moser, J. Med. Chem., 2008, 51, 2871 CrossRef PubMed.
- B. L. Angus, A. M. Carey, D. A. Caron, A. M. Kropinski and R. E. Hancock, Antimicrob. Agents Chemother., 1982, 21, 299 CrossRef CAS.
- C. K. Maurer, A. Steinbach and R. W. Hartmann, J. Pharm. Biomed. Anal., 2013, 86, 127 CrossRef CAS PubMed.
- F. Lépine, F. E. Déziel, S. Milot and L. G. Rahme, Biochim. Biophys. Acta, 2003, 1622, 36 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis and analytics of all synthetic intermediates and all substrates used in the enzyme inhibition assay as well as procedure of the mutagenicity test; HHQ and PQS inhibition in the PA14 wild-type strain. See DOI: 10.1039/c4ob00707g |
| This journal is © The Royal Society of Chemistry 2014 |