Open Access Article
Open Access ArticleNanofluidics in point of care applications
L. I.
Segerink
* and
J. C. T.
Eijkel
*
BIOS Lab on a Chip Group, MESA+ Institute for Nanotechnology, University of Twente, Enschede, the Netherlands. E-mail: l.i.segerink@utwente.nl; j.c.t.eijkel@utwente.nl
First published on 15th May 2014
Abstract
Nanofluidics is generally described as the study of liquid flow in or around structures of 100 nm or smaller, and its use for lab on a chip devices has now been actively studied for two decades. Here a brief review is given of the impact that this nanofluidics research has had on point of care applications. Four areas are identified where nanofluidics has brought the largest contributions: single nanopores, nanoporous membranes, nanoconfinement and the use of concentration polarization. The sometimes revolutionary developments in these areas are briefly treated and finally challenges and future perspectives are described.
Introduction
It is important to clearly outline the area which we will discuss in this insight paper. Here we will focus on point of care (POC) applications of nanofluidics proper, hence involving the flow of liquid around or in structures of the order of 100 nm or smaller. Thus we will not treat POC applications based on the use of sensing nanostructures such as quantum dots, nanobeads, plasmonic nanostructures or nanowires. The subject thus properly will be ‘nanofluidics in POC applications’ and not ‘nanotechnology in POC applications’. As a general remark we want to repeat Whitesides' dictum that for everything there is a right size. Thus smaller is not (always) better. The use of microfluidics for POC can be more relevant for certain applications than using nanoscale devices.1Subsequently it is important to stress already at the start of this paper that this area has a quite interesting split development. On the one side there are the applications for DNA sequencing in which much nanofluidics is involved, which commercially race ahead at such a pace that POC applications of these technologies can be expected in the not-too-far future. On the other side there are the applications for e.g. protein sensing (such as HIV diagnostics) that develop at a much slower pace but are generically directly aimed at POC use. It is quite probable that (parts of) the technology developed in the first area will later be of use in the second area. In view of this, we will discuss both developments in parallel in the following treatment.
In the remainder of this review we will let ourselves be guided by a phenomenological subdivision, distinguishing a number of different nanofluidic phenomena important for POC applications and treating the related developments towards POC applications.
State-of-the-art
Nanofluidics is generally described as the study of liquid flow in or around structures of 100 nm or smaller. At this scale the system behaviour is strongly determined by the forces between wall and solvent and wall and solute molecules, which can be electrostatic, van der Waals, hydrophobic or entropic forces.2 During transport furthermore frictional forces (slip or no-slip conditions) are important. Finally, Born repulsion between atoms plays an important role preventing solutes to enter very small structures.Nanofluidics is roughly applied in four different ways for analysis: by using single nanopore transport, nanoporous membranes, nanoconfinement, and by the concentration polarization functionality. When single nanopores or nanoporous membranes are used for analysis, transport of analyte, solvent and solutes will simultaneously take place. In this case the analyte confinement in the pore plays an important role (as in the case of the cavities) but also the various interactions between solutes, solvent and pore wall during transport. Nanofluidic confinement in cavities or droplets allows very sensitive analysis because the ultrasmall measurement volume gives a low background signal (e.g. light emission) and because small amounts of product cause high concentration changes. Finally, nanofluidic structures can also be used as functional device elements for sample treatment. Especially the phenomenon of concentration polarization is then used to increase or decrease the analyte concentration as well as the concentrations of other (ionic) solutes.
Single nanopores
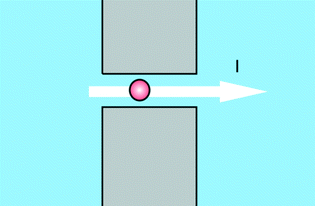
An important group of applications centres on the use of structures as depicted in Fig. 1. Using this structure a measurement can be performed which can be seen as a variation of the Coulter counter principle, where a current across a pore is modulated by the passage of analytes. While Coulter counters use micropores to detect micron-size particles such as biological cells, the use of nanopores enables the detection of nano-size particles such as viruses, protein molecules or nucleic acid molecules.3 This type of sensors allows for high-throughput, single molecule detection and characterization and can be integrated into microfluidic systems for sample routing. Added advantages are, that information on many single molecules is obtained, and that interactions of these molecules with e.g. custom-made ligands can simultaneously be detected. The versatility of the nanopore concept is underlined by the fact that it has also been proposed for detection of life on other planets, since it is not necessarily restricted to the detection of the genetic information carrier DNA, but in general can be applied to sequence chain-like genetic information carriers without knowing the exact building blocks of it.4The furthest technological advances of this type of device have recently been booked in companies, outside of the academic environment, developing nanopore DNA sequencing. An overview of the latest developments was given by Schneider and Dekker.5 The most promising research has focused on using α-hemolysin in a lipid bilayer as the pore. Sequencing is then performed by binding the DNA to a polymerase and slowly unzipping it while feeding the single strand through the pore (Oxford Nanopore Technologies), or extending a single strand DNA by a polymerase tethered to the pore and measuring the current changes through the pore when specific nucleotide tags are released during the polymerase reaction (Genia). Though not all hurdles for commercialization have been crossed, nanopore sequencing is one of the promising next-next generation sequencing methods due to its potential infinite read lengths and single molecule capabilities.6 As we already signalled in the introduction of this paper, the powerful technology developed for DNA sequencing can also be applied for POC measurements. Oxford Nanopore Technologies for example on its website announces a disposable device for single molecule sensing in biological fluids.
In academia, large strides have been made in the past ten years in the development of nanopore protein sensing, especially in changing the pore surface and adding functionalities such as specific ligands for the analytes of choice, or detecting both the passage of the free analyte and its antibody-bound form.7,8 In general these sensors will not be single molecule sensors but will measure an averaged transport rate.
Nanoporous membranes
Nanoporous membranes result from placing multiple nanopores in parallel, and exhibit a much larger transport flux. Also nanoslits, which have one dimension on the nanoscale, effectively function as many parallel nanopores. The passage of substances through such membranes or slits, just as in the case of single nanopores, can be tuned by the various filtering mechanisms such as electrostatics, molecular size or entropic energy. For protein sensing, nanoslits can be used functionalized with specific ligands, and the passage of current recorded to detect ligand binding. In this case the measured currents are advantageously larger than with single nanopores.9Nanoconfinement
Nanofluidics is often applied for measurements where confinement in ultrasmall volumes (nanocavities) is of prime importance (Fig. 2). The specific advantage of small volumes is, that the background signal (e.g. fluorescence emission) is small, and that the product (e.g. of an enzyme reaction) is only little diluted. Nanocavities are for example used for DNA sequencing, and devices with nanocavities are already commercially available. Based on the pioneering work of Webb and Craighead,10 Pacific Biosciences has developed a platform with massively parallel nanofluidic cavities coupled to zero mode waveguides, which are used to optically observe the addition of single nucleotides to a single stranded DNA molecule immobilized in the cavity. Confinement here is used to precisely determine the location of the reaction (and hence the location of the fluorescent product) in the illuminated volume. Also in the sequencing technology of Ion Torrent confinement plays a central role. Here a bead with multiple identical single stranded DNA strands is placed in a nanofluidic cavity on top of a CMOS ISFET. When a single nucleotide extension reaction takes place, the liberated protons are sensed by the ISFET. The CMOS architecture allows massive parallelization and convenient readout. It can be envisaged that the rapidly reducing cost and analysis time of these devices in time will make them part of the POC arsenal. Also, applications of these powerful technologies for e.g. protein sensing seem probable.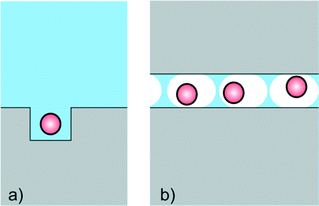 | ||
| Fig. 2 Cartoon for devices based on the use of nanoconfinement by a) nanocavities or b) femtoliter emulsion droplets. | ||
Nanogaps have been used in combination with FET-type sensors to detect biomolecules in a label-free way.11 The nanogap allows the precise spatial location of the sensing molecules with respect to the sensor. It is situated inside the gate material and filled with molecules which bind specifically to the biomolecule of interest. Attachment of the biomolecule will change the dielectric constant and the capacitance of the gate, resulting in a change of the threshold voltage which can be measured.11 Such a configuration can be used to measure antigens for Avian influenza, although it is not yet demonstrated in clinical samples.12 Additionally the detection of C-reactive protein in human serum was shown.13 Although multiple sensors were needed to retrieve a reliable signal, protein detection limits as low as 0.1 ng mL−1 were demonstrated.13
Another very versatile form of nanoconfinement is in emulsion droplets. The use of an emulsion prevents diffusion and hence light emission can be measured against a very low (integrated) background. Recently a single molecule-counting immunoassay was demonstrated in femtoliter droplets, opening the way for the development of highly sensitive immunoassays.14 In this context also digital PCR is worth mentioning, which uses picoliter droplets in which after dilution single DNA molecules are amplified.15,16 Digital PCR reduces the amount of cycles needed thereby making faster analysis possible,15 and can also be used to identify specific sequences in an isolated single molecule.17 Digital PCR is a good example of the advantages of confinement. At the same time it doesn't benefit from downscaling further than to micrometer-diameter droplets, thereby demonstrating the concept of ‘the right size’.
Nanoconfinement has also been used for highly sensitive electrochemical detection by redox cycling, using integrated electrodes at opposing sides of a nanocavity or nanochannel. Single molecule detection capability was demonstrated, in first instance in an organic solution,18 and later also in an aqueous solution.19 Until now only model redox compounds were detected with this device, however since physiological salt concentrations can be used this method is a promising tool for single molecule detection in biological fluids.19
Nanoconfinement in nanochannels can also be used in combination with optical detection techniques to detect individual human viruses and bacteriophages in fluids. The passage of these particles through a nanochannel causes a change in optical signal which depends on the particle's diameter. Using this method the concentration of different sizes of nanoparticles can be determined, such as for instance the HIV virus concentration.20
Genome mapping is another field that can benefit from structures for nanoconfinement. DNA strands can be stretched in nanochannels21 for subsequent analysis. An example of such an analysis is restriction mapping whereby DNA molecules are cut at specific locations by restriction enzymes. In this way a restriction map with 1.5 kbp resolution can be obtained within 60 seconds.22 In another approach specific sequences on elongated and confined DNA in a nanochannel are imaged by inserting fluorescent nucleotides at the specific sequences on one of the strands.23 In these two examples, enzymes are needed to map the DNA, while this can be avoided when a denaturation map is made. In denaturation mapping, the temperature of a nanoconfined DNA molecule is raised, causing specific parts of the molecule to melt, depending on the local abundance of adenine or thymine. The resulting melting pattern can be optically measured, since the fluorescently labelled DNA will show less fluorescence at the denatured regions.24 The method can be used as a tool to investigate the entire genome of single cells.24 It can be envisioned, that such rapid DNA mapping techniques in time find application in healthcare diagnostics at the point of care, e.g. for cancer cell heterogeneity determination.
Concentration polarization
When a membrane, filter or nanochannel blocks or reduces the transport of a certain substance, this substance is accumulated at the feed side of the structure and depleted at the drain side. This quite general phenomenon is called concentration polarization. In a special case of great practical relevance, an electrical field is applied for transmembrane transport and the transported substance is ionic. In that case the requirement of bulk electroneutrality will also cause changes in the concentration of the counterions, and as a result salt depletion and accumulation will occur at the opposite sides of the membrane (Fig. 3) accompanied by changes in the local electrical conductivity. In Lab on a Chip devices many types of structures (frits, nanochannels, Nafion membranes, charged gels) have been used to generate salt concentration polarization. In POC applications, nanofluidic functional units can be integrated to achieve concentration polarization and be used for various purposes. Ionic analyte will generally accumulate at the start of the depletion zone, and this accumulation can then be used for example to increase the sensitivity of binding assays such as immunoassays.25,26 Also, the decrease of salt concentration can be used to increase the sensitivity of potentiometric (e.g. nanowire) assays.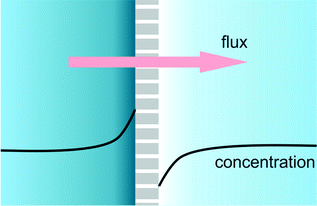 | ||
| Fig. 3 Cartoon of a concentration polarization functional unit, based on selective transport through nanochannels, nanopores, gels, frits or membranes. | ||
Challenges and future perspectives
Some nanofluidics applications are already fully commercialized (Ion Torrent), some almost (Oxford Nanopore technologies), and some are still in the academic research phase. In each different phase the challenges are of a different nature. POC systems are most useful when ‘ASSURED’: Affordable, Sensitive, Specific, User-friendly, Rapid and Robust, Equipment-free, Deliverable.27 Of these criteria, cheap, accurate and rapid/robust are the most critical. As shown here, the use of nanofluidics has enabled a number of previously impossible sample manipulation and detection methodologies, which have greatly improved system sensitivity and speed. Price will come down with increasing production volume and the availability of more sophisticated techniques. The $1000 genome is practically there, bringing this technique almost to the standard clinical laboratory, and it is to be envisioned that in the future a genome analysis will be made for every neonate.Nanofluidics enables some very sophisticated techniques, some of which are very useful for POC applications and have already been demonstrated in academic research. Perhaps the biggest remaining challenge for the nanofluidics field is to bridge the gap for the techniques mentioned in this paper to bring them from the academic lab to the commercial application. This is a valorisation challenge, which with different degrees of success is confronted throughout the world.
Notes and references
- G. M. Whitesides, Nat. Biotechnol., 2003, 21, 1161–1165 CrossRef CAS PubMed.
- J. C. T. Eijkel and A. van den Berg, Microfluid. Nanofluid., 2005, 1, 249–267 CrossRef CAS.
- Z. Siwy, L. Trofin, P. Kohli, L. A. Baker, C. Trautmann and C. R. Martin, J. Am. Chem. Soc., 2005, 127, 5000–5001 CrossRef CAS PubMed.
- F. Rezzonico, Astrobiology, 2014, 14, 344–351 CrossRef CAS PubMed.
- G. F. Schneider and C. Dekker, Nat. Biotechnol., 2012, 30, 326–328 CrossRef CAS PubMed.
- S. Kumar, C. J. Tao, M. C. Chien, B. Hellner, A. Balijepalli, J. W. F. Robertson, Z. M. Li, J. J. Russo, J. E. Reiner, J. J. Kasianowicz and J. Y. Ju, Sci. Rep., 2012, 2, 684 Search PubMed.
- S. Howorka and Z. Siwy, Chem. Soc. Rev., 2009, 38, 2360–2384 RSC.
- K. J. Freedman, A. R. Bastian, I. Chaiken and M. J. Kim, Small, 2013, 9, 750–759 CrossRef CAS PubMed.
- N. F. Y. Durand and P. Renaud, Lab Chip, 2009, 9, 319–324 RSC.
- M. J. Levene, J. Korlach, S. W. Turner, M. Foquet, H. G. Craighead and W. W. Webb, Science, 2003, 299, 682–686 CrossRef CAS PubMed.
- H. S. Im, X. J. Huang, B. Gu and Y. K. Choi, Nat. Nanotechnol., 2007, 2, 430–434 CrossRef CAS PubMed.
- B. Gu, T. J. Park, J. H. Ahn, X. J. Huang, S. Y. Lee and Y. K. Choi, Small, 2009, 5, 2407–2412 CrossRef CAS PubMed.
- C. H. Kim, J. H. Ahn, J. Y. Kim, J. M. Choi, K. C. Lim, T. J. Park, N. S. Heo, H. G. Lee, J. W. Kim and Y. K. Choi, Biosens. Bioelectron., 2013, 41, 322–327 CrossRef CAS PubMed.
- J. U. Shim, R. T. Ranasinghe, C. A. Smith, S. M. Ibrahim, F. Hollfelder, W. T. S. Huck, D. Klenerman and C. Abell, ACS Nano, 2013, 7, 5955–5964 CrossRef CAS PubMed.
- N. R. Beer, B. J. Hindson, E. K. Wheeler, S. B. Hall, K. A. Rose, I. M. Kennedy and B. W. Colston, Anal. Chem., 2007, 79, 8471–8475 CrossRef CAS PubMed.
- B. J. Hindson, K. D. Ness, D. A. Masquelier, P. Belgrader, N. J. Heredia, A. J. Makarewicz, I. J. Bright, M. Y. Lucero, A. L. Hiddessen, T. C. Legler, T. K. Kitano, M. R. Hodel, J. F. Petersen, P. W. Wyatt, E. R. Steenblock, P. H. Shah, L. J. Bousse, C. B. Troup, J. C. Mellen, D. K. Wittmann, N. G. Erndt, T. H. Cauley, R. T. Koehler, A. P. So, S. Dube, K. A. Rose, L. Montesclaros, S. L. Wang, D. P. Stumbo, S. P. Hodges, S. Romine, F. P. Milanovich, H. E. White, J. F. Regan, G. A. Karlin-Neumann, C. M. Hindson, S. Saxonov and B. W. Colston, Anal. Chem., 2011, 83, 8604–8610 CrossRef CAS PubMed.
- B. Vogelstein and K. W. Kinzler, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9236–9241 CrossRef CAS.
- M. A. G. Zevenbergen, P. S. Singh, E. D. Goluch, B. L. Wolfrum and S. G. Lemay, Nano Lett., 2011, 11, 2881–2886 CrossRef CAS PubMed.
- S. Kang, A. Nieuwenhuis, K. Mathwig, D. Mampallil and S. G. Lemay, ACS Nano, 2013, 7, 10931–10937 CrossRef CAS PubMed.
- A. Mitra, F. Ignatovich and L. Novotny, Biosens. Bioelectron., 2012, 31, 499–504 CrossRef CAS PubMed.
- J. O. Tegenfeldt, C. Prinz, H. Cao, S. Chou, W. W. Reisner, R. Riehn, Y. M. Wang, E. C. Cox, J. C. Sturm, P. Silberzan and R. H. Austin, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10979–10983 CrossRef CAS PubMed.
- R. Riehn, M. C. Lu, Y. M. Wang, S. F. Lim, E. C. Cox and R. H. Austin, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10012–10016 CrossRef CAS PubMed.
- K. Jo, D. M. Dhingra, T. Odijk, J. J. de Pablo, M. D. Graham, R. Runnheim, D. Forrest and D. C. Schwartz, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2673–2678 CrossRef CAS PubMed.
- W. Reisner, N. B. Larsen, A. Silahtaroglu, A. Kristensen, N. Tommerup, J. O. Tegenfeldt and H. Flyvbjerg, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 13294–13299 CrossRef CAS PubMed.
- Y. C. Wang and J. Y. Han, Lab Chip, 2008, 8, 392–394 RSC.
- S. H. Ko, S. J. Kim, L. F. Cheow, L. D. Li, K. H. Kang and J. Han, Lab Chip, 2011, 11, 1351–1358 RSC.
- R. W. Peeling and D. Mabey, Clin. Microbiol. Infect., 2010, 16, 1062–1069 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |