Research highlights
Gulden
Camci-Unal
ab,
Šeila
Selimović
ab,
Mehmet R.
Dokmeci
ab and
Ali
Khademhosseini
*abcd
aCenter for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Cambridge, Massachusetts 02139, USA. E-mail: alik@rics.bwh.harvard.edu
bHarvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA
cWyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts 02115, USA
dWorld Premier International – Advanced Institute for Materials Research (WPI-AIMR), Tohoku University, Sendai 980-8577, Japan
First published on 23rd January 2013
Biomimetic honeycombs
Honeycombs consist of hexagonal unit cells and are commonly used for storage of honey, pollen, or larvae by bees.1 The honeycomb geometry offers the tightest packing of materials with the least amount of material.2 Using this geometry in an industrial setting can significantly reduce the cost, density and mass of the product while enhancing its mechanical properties. It may be possible to develop novel, composite materials for aerospace, packaging, environmental catalyst and electronics industries by mimicking the geometry of natural honeycombs.3 Based on its resemblance to natural material structures, honeycomb geometry can also be used in tissue engineering applications. Although this geometry is advantageous for many reasons, the existing methods to fabricate honeycomb-mimetics have some significant drawbacks. For example, current methods require complex instrumentation and protocols and do not yield robust and reliable hexagonal structures.4To tackle these issues, Qin and colleagues have recently synthesized honeycomb structures by a double emulsion templating strategy.4 They fabricated nanoporous poly(lactic-co-glycolic acid) (PLGA) micro/nanoparticles by polymer precipitation and salt leaching. Unlike the existing methods, this simple approach has enabled generation of lightweight, densely packed and mechanically strong structures similar to the native honeycomb geometry. The resulting PLGA microspheres with honeycomb structures could potentially be used as efficient functional drug carriers.
In this work, a double emulsion approach was utilized by flow focusing in a poly(dimethylsiloxane) (PDMS) microfluidic device. The double emulsion phases were generated using ammonium bicarbonate (NH4HCO3) as the inner phase, oil/PLGA as the middle phase, and poly(vinyl alcohol) (PVA) as the outer phase during the flow focusing process. The emulsification via this configuration resulted in nanoporous membranes on the PLGA microspheres (Fig. 1). The unique hexagonal shapes of the honeycomb structures were highly influenced by the use of a low boiling point organic solvent, namely dimethyl carbonate. Its rapid evaporation during the PLGA microsphere formation resulted in generation of the honeycomb geometry. The average diameter (50–100 μm) of the microspheres was influenced by the concentration of NH4HCO3 in the inner phase of the double emulsion.
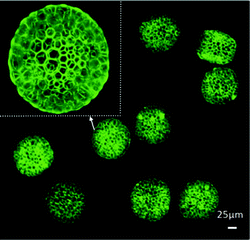 | ||
| Fig. 1 Confocal images of PLGA microspheres with honeycomb features. Figure reprinted with permission from Ma et al.4 | ||
It was found that the biomimetic PLGA microspheres possessed high membrane permeability enabling selective release of small molecules, such as drugs and proteins. Mesenchymal stem cells (MSCs) adhered, spread, and proliferated on the honeycomb microspheres for at least 10 days facilitated by the oxygen- and nutrient-permeable nanoporous membrane. Also, the effect of the release of an encapsulated drug, carboplatin, from the microspheres on HepG2 cells was studied by testing the apoptosis and growth inhibition behavior after 3 days of culture. As expected, it was found that carboplatin-loaded honeycomb microspheres were able to suppress tumor activity. Unlike existing strategies, this approach allows for encapsulation of not only hydrophilic drugs, but also their hydrophobic analogs.
The PLGA microspheres with unique honeycomb structures present a novel approach to mimic natural materials. The synthesis method presented here is simple and straightforward and it stands out from other approaches due to the unique combination of double emulsion templating, PLGA precipitation and internal salt leaching process. The resulting microspheres were found to be cytocompatible, opening up new potential applications in biomedical research. For example, the microspheres could be useful for delivering drugs to tissues for repair or to direct stem cell differentiation. The functionalized microspheres could also be used in other areas of biology as well as environmental catalysis suggesting new research avenues.
Microfluidic vessel chips
Mechanical stress, strain, and shear influence the differentiation behavior of stem cells and cellular function.5 For example, cardiovascular tissues are continuously mechanically stimulated by the pulsatile blood flow.6 This may be relevant to vascular development, disease pathogenesis, and tissue formation or remodeling. However, it is difficult to perform long-term and high-throughput cell culture experiments using existing techniques. Systems that can mimic the mechanical cues for small blood vessels are also difficult to implement. Thus, there is a tremendous need to develop in vitro systems that can address these limitations.Zhou et al. have recently designed and developed a microfluidic chip to provide cyclic radial strain by hydrodynamic actuation for human MSCs within an array of channels ranging from 20 to 500 μm in width.6 This was enabled by separating the cell culture wells from the pressurized channels with an elastic membrane (Fig. 2). In this study, different channel widths allowed for simultaneous application of different mechanical strains. In the body, arteries stretch radially under pulsatile blood flow. Therefore, this microfluidic setup was utilized to closely mimic native conditions.
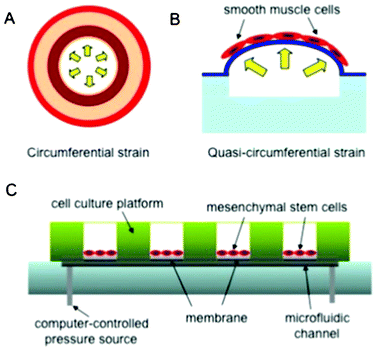 | ||
| Fig. 2 Schematic of the microfluidic chip, inspired by Zhou et al.6 (A) Circumferential strain in native arteries. (B) Quasi-circumferential strain in the microfluidic vessels. (C) Schematic of the assembled microfluidic set-up. | ||
The channels were covered with deformable membranes, where the human MSCs were cultured for up to 7 days with high cellular viability. Cyclic stretch was applied to the cell cultures for 6 days without fatigue or decrease in the performance of the fluidic device. This was reported as the longest in vitro cell culture with continuous cyclic stimuli. Circumferential strain values up to 20% were used for mechanical stimulation to mimic the mechanical conditions in the native arteries. It was noted that the cellular alignment and localization of MSCs were significantly improved after exposure to strains above 10%. This suggests that dynamic mechanical stimulation is a critical component to control cellular behavior in small blood vessels. In addition to viability and alignment analyses, the transduction of cell signaling in response to dynamic mechanical loading was also probed. The signaling proteins that are responsible for inducing differentiation or maintaining the undifferentiated state (GSK-3β, β-catenin/Phospho-β-catenin, Phospho-SMAD1 and SMAD2/Phospho-SMAD2) were simultaneously detected within the microfluidic chip by performing immunofluorescence staining and quantification of fluorescence intensity.
The reported strategy allows for rapid screening of cellular behavior in response to mechanical stimuli. The microscale biomimetic platform enables high-throughput monitoring of cellular alignment or differentiation upon cyclic strain and is able to mimic the physiological conditions in small blood vessels. The microfluidic chip is robust, versatile, and suitable for long term dynamic stimulation of cell cultures. This strategy could be used in regenerative engineering where mechanical stimulation is important to control cellular behavior, fate, and function as well as development of disease states and tissue formation.
A new guide for teaching BioMEMS
The field of BioMEMS (Bio-microelectromechanical systems) combines principles of physics, mechanical and electrical engineering to enable microscale studies of biological systems, such as stem cells or the formation of tissues in vitro. Perhaps due to the rapid progress in the field, most relevant information is being disseminated at conferences and through research articles. Lately, review papers covering a specific problem in detail or books offering a broad outlook on the field have added to the relevant literature.7–9 However, if BioMEMS is to become a regular part of engineering education, then a textbook covering the field in depth and breadth would also be needed.Dr Albert Folch of University of Washington has recently published just such a book, entitled “Introduction to BioMEMS”10 based on his popular university course (Fig. 3). The textbook is geared towards science and engineering students at the undergraduate and graduate levels. Intended to describe the importance of microscale research platforms for biological and medical research, the text offers a clear image of the development of BioMEMS by reviewing seminal, high-impact innovations and advancements in the field in comparison with traditional alternatives.
 | ||
| Fig. 3 The introductory textbook on MEMS for biological applications contains over 400 figures and exercises, and is accompanied by a set of free downloadable powerpoint slides for instruction.10 | ||
The reader is introduced to the field of BioMEMS with a justification for microscale biological research, as compared to traditional, macroscale laboratory methods. These, of course, include the reduced reagent amounts, lower cost, and improved control over various experimental parameters. The author then covers a range of fabrication and micropatterning techniques for use with both MEMS and biological materials (proteins and cells), since knowledge of various device fabrication methods is a prerequisite for functional chip design. A section centering on hydrogel devices exemplifies the importance of newly developed biomaterials in biomedical research.
An entire chapter of the book is devoted to microfluidic technologies, covering the basics of fluid dynamics inside small channels, operation of microfluidic platforms and device elements such as mixers and multiplexers. The author also discusses different types of droplet-based microfluidics such as electrowetting or digital platforms as well as air and oil carrier platforms and their uses. Importantly, throughout this chapter Folch mentions several common difficulties in operating microfluidic devices, such as formation of air bubbles that can destabilize the flow and in certain cases contribute to cell death. Although certain topics including particle flow and substrate tension could have been explored in more detail, the included information and references are well suited for an introductory book on BioMEMS.
The second part of the textbook focuses on biological applications of miniaturized devices. The uninitiated reader benefits from a detailed discussion of potential experimental difficulties and solutions relating to sample manipulation. A comprehensive section on conducting molecular biology experiments on a microfluidic chip introduces a range of immunoassay-based devices, including pregnancy tests and devices suitable for screening of infectious diseases. Miniaturized devices that enable gene expression in a high-throughput manner are also presented and reviewed in the context of the modern fields of genomics and proteomics.
Helpful and interesting for new and experienced BioMEMS researchers, the chapter on BioMEMS for cell biology covers both the achievements of the field (e.g. in cell–substrate and cell–cell signaling) as well as the limitations. Here, a particular focus is on chips for cellular neurobiology, including devices that can measure the electrical activity of neurons or help us uncover the biology behind the sense of smell. The following tissue engineering chapter describes approaches for generating miniature living tissue in vitro. The textbook that began with a justification for BioMEMS ends with one of the ultimate goals of the field – developing implantable microscale devices for diagnostics and therapy.
In this classroom-tested textbook, Folch offers a broad, yet detailed lecture on BioMEMS, sharing information in clear, straight-forward language and over 400 excellent color figures. The inclusion of only the most important formulae and thought experiments instead of involved mathematical exercises make this textbook easily readable and relevant both to entry level as well as advanced researchers. This also makes “Introduction to BioMEMS” a perfect guide and an easy-to-use comprehensive textbook for researchers from different science and engineering fields. In addition, for instructors designing a new course focusing on BioMEMS, powerpoint slides accompanying the textbook are made available through http://faculty.washington.edu/afolch/TextbookMap.html.
References
- K. Zhang, H. L. Duan, B. L. Karihaloo and J. X. Wang, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 9502–9506 CrossRef CAS.
- C. M. Taylor, C. W. Smith, W. Miller and K. E. Evans, Int. J. Solids Struct., 2011, 48, 1330–1339 CrossRef.
- J. Wang, H. X. Shen, C. F. Wang and S. Chen, J. Mater. Chem., 2012, 22, 4089–4096 RSC.
- J. Ma, Y. S. Hui, M. Zhang, Y. Yu, W. Wen and J. Qin, Small, 2012 DOI:10.1002/smll.201201624.
- D. J. Wang, J. S. Park, J. S. F. Chu, A. Krakowski, K. X. Luo, D. J. Chen and S. Li, J. Biol. Chem., 2004, 279, 43725–43734 CrossRef CAS.
- J. Zhou and L. E. Niklason, Integr. Biol., 2012, 4, 1487–1497 RSC.
- A. C. R. Grayson, R. S. Shawgo, A. M. Johnson, N. T. Flynn, L. Yawen, M. J. Cima and R. Langer, Proc. IEEE, 2004, 92, 6–21 CrossRef CAS.
- E. Nuxoll and R. Siegel, IEEE Eng. Med. Biol. Mag., 2009, 28, 31–39 CrossRef.
- S. S. Saliterman, Fundamentals of BioMEMS and Medical Microdevices, SPIE Press, 2006 Search PubMed.
- A. Folch, Introduction to BioMEMS, CRC Press, 2013 Search PubMed.
| This journal is © The Royal Society of Chemistry 2013 |
