DNA origami technology for biomaterials applications
Masayuki
Endo
*ab,
Yangyang
Yang
c and
Hiroshi
Sugiyama
*abc
aInstitute for Integrated Cell-Material Sciences (WPI-iCeMS), Kyoto University, Yoshida-ushinomiyacho, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: endo@kuchem.kyoto-u.ac.jp; hs@kuchem.kyoto-u.ac.jp; Fax: +81-75-753-3670
bCREST, Japan Science and Technology Corporation (JST), Sanbancho, Chiyoda-ku, Tokyo 102-0075, Japan
cDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa-oiwakecho, Sakyo-ku, Kyoto 606-8502, Japan
First published on 7th December 2012
Abstract
DNA origami is an emerging technology for designing and constructing defined multidimensional nanostructures. This technology is now expanding to materials science. This article introduces the basics of DNA origami, the design of various two-dimensional and three-dimensional DNA origami structures, and the programmed assembly of origami structures. DNA origami has unique properties, such as an addressable surface, which enables selective functionalization with biomolecules and nanomaterials. The origami can also be combined with top-down nanotechnology, such as placement on a fabricated substrate. The technology is also applied to single-molecule imaging and analysis systems constructed on designed DNA origami structures. Furthermore, DNA mechanical nanodevices working on DNA origami have been realized, and cell-oriented applications are now in progress. DNA origami technology has practical potential in various research fields.
1. Introduction
DNA materials science related to structural DNA nanotechnology has grown rapidly as a research area in the past two decades. The technology allows the construction of various self-assembled scaffolds, which can be used for the placement and arrangement of functional molecules and nanomaterials and for the production of complex molecular devices. The field of DNA nanotechnology was pioneered by Ned Seeman, who created various important DNA motifs and strategies for self-assembly which constitute the basic concept of structural DNA nanotechnology.1,2 DNA nanotechnology, which is now applied for the construction of nanoscale structures and functionalized materials, is further used in molecular mechanics and computation and in the fields of synthetic chemistry and biology, and it continues to develop in response to technology demands.3–5 “DNA origami”, a new programmed DNA assembly system based on well-established DNA nanotechnology, enables the design of two-dimensional (2D) nanostructures with a wide variety of shapes of defined size.6 Moreover, functional molecules and nanoparticles have been placed on DNA origami structures.3–5This article highlights the latest research related to origami-based DNA nanotechnology and describes the expansion of the DNA origami method to biomaterial applications. This review focuses on the basics of this technology, including the design and construction of various 2D and three-dimensional (3D) structures and programmed arrangements, and describes its application in the selective functionalization and single-molecule imaging of biomolecules, cell-targeted applications, and molecular machines built on a DNA origami scaffold.
2. 2D DNA origami
DNA origami, developed by Rothemund in 2006, has enabled the construction of a wide variety of 2D structures of around 100 nm in size, including rectangles, triangles, and even a smiley face and five-pointed star (Fig. 1).6 In this method, a long single-stranded DNA (M13mp18; 7249 nucleotides) and the sequence-designed complementary strands (called “staple strands”; most are 32-mer) are mixed and then annealed from 95 °C to room temperature over 2 h, resulting in the formation of target structures by self-assembly (Fig. 1A). The structure can be imaged using atomic force microscopy (AFM), and the assembled structure can be formed according to a design. To create 2D DNA origami structures, adjacent double-stranded DNAs (dsDNAs) should be connected to each other via a crossover. In this design, the geometry of the double helices involved has three helical rotations for 32 base pairs (Fig. 1B). For example, two neighboring crossovers of the central dsDNA in an arrangement of three adjacent dsDNAs should be located at the opposite sites (180° rotated, 0.5 turns). Therefore, the crossovers should be separated by 16 base pairs (1.5 turns). To maintain a stable planar structure, this rule should be followed when placing multiple staple strands. DNA origami structures are formed using many different staple strands, so that hairpin DNA markers can be placed at any position on the surface of the DNA structure. Hairpin DNA used as a topological marker is observed as a dot under AFM imaging. In this case, hairpins are placed perpendicular to the surface of the origami, therefore each hairpin should be placed at a position eight base pairs from the crossover (270° rotation). The distance between the centers of the adjacent staples is about 6 nm, so the adjacent hairpins can be observed as distinct spots according to the spatial resolution of the AFM. Using the hairpin markers, patterns, such as the map of a hemisphere (Fig. 1D), can be drawn precisely on the DNA origami surface.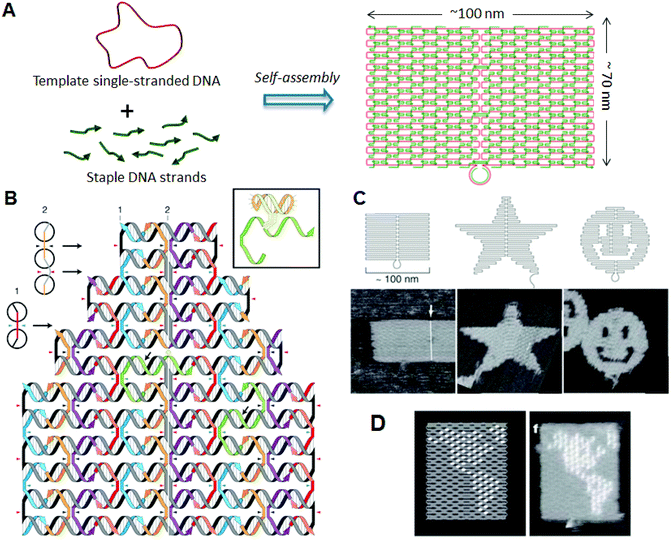 | ||
| Fig. 1 DNA origami structure. (A) The method employed to prepare a DNA origami structure from the template single-stranded DNA and staple strands. (B) Design of a self-assembled DNA origami structure and geometry of the incorporated dsDNAs. Colored strands and a gray/black strand represent staple strands and template single-stranded DNA, respectively. Staple strands connect the adjacent duplexes with crossovers. Inset: Structure of hairpin DNA for a topological marker. (C) Design and AFM images of self-assembled DNA origami structures. (D) Drawing of a hemisphere on the DNA origami with hairpin DNAs (white dots) and an AFM image of the assembled DNA origami. | ||
In addition, when functional molecules and nanoparticles are conjugated to specific staples, they can be placed on the origami surface at selected sites. 2D structures formed with the DNA origami system not only have shape variations—a remarkable property of DNA origami is that all the positions of the structure have DNA sequence information (an address). Before the emergence of DNA origami technology, it was difficult to create ∼100 nm-sized structures through the self-assembly of small DNA components. DNA origami technology solved this problem and provided the breakthrough that enabled expansion of the shape design and the creation of addressable structures.
Since the DNA origami system uses a long single-stranded template DNA, the size of the 2D structure is determined by the length of the template strand. Various single-stranded DNAs were isolated for use as a template in the preparation of DNA origami.7,8 A strategy of using DNA tiles (17 × 16 nm) instead of staples has also been developed, which allows size expansion by the introduction of 25 to 56 DNA tiles.9
3. Programmed arrangement of multiple DNA origami components
The programmed arrangement of DNA origami is an important technique for preparing the desired large structures, particularly in terms of integrating complicated functions. We explored techniques for arranging multiple DNA origami components, and developed methods to arrange rectangular DNA origami tiles horizontally in a programmed fashion.10 As the ends of the helical axes align at both edges of the DNA origami rectangles, a rectangular DNA origami tiles horizontally in a programmed fashion.10 In addition, as the ends of the helical axes align at both edges of the DNA origami rectangles, rectangular origami tiles assemble via π-interaction at the edges. We introduced specific concave and convex connectors into the origami rectangles to align these rectangles precisely with neighboring origami tiles. DNA rectangles should correctly assemble by shape and sequence complementarity, where complementary strands are introduced into the concavity and the convex connectors. After self-assembly, we observed that the DNA tiles were aligned and oriented in the same direction. Furthermore, to align origami tiles accurately, the positions of the connectors and the concavity were changed to connect two specific tiles. Five tiles were designed to align horizontally. In this system, we adopted a two-step self-assembly: first, individual origami tiles were prepared, and then the multiple tiles were assembled in the second stage by slower annealing from 50 °C. The DNA origami is stable enough to heat at 50 °C in secondary annealing. To allow identification of the DNA tiles, hairpin markers were introduced onto the individual tiles. After self-assembly, judging by the order of the markers, the five tiles were aligned correctly. In addition, hairpin markers were used to display letters of the alphabet on the origami surface. The letters D, N, A, N, and O were each introduced onto one of the five tiles. After self-assembly using the first three tiles and last four tiles, the words “DNA” and “NANO” were displayed, respectively. Using five tiles with the letters K, Y, O, T, and O, the five-letter word “KYOTO” was formed. We named these designed DNA tiles “DNA jigsaw pieces”.The method described above was applied to prepare the 2D assembly system.11 The shape and sequence selectivity were introduced to both lateral edges for extension in the vertical direction (Fig. 2A). Nine DNA tiles were designed and prepared. Three tiles were then programmed to be connected vertically or horizontally, and three sets of vertical or horizontal trimers were finally assembled into a 3 × 3 assembly with ∼30% yield; this assembly was confirmed by hairpin markers introduced onto the individual origami tiles. Taking a different approach, we explored novel 2D assemblies. Four connection sites on the four-way DNA origami connector were oriented to the exterior to facilitate connection between the edges of neighboring DNA jigsaw tiles via π-interaction.12 Using this four-way connector, five and eight origami monomers were assembled to form a cruciform and a hollow square structure, respectively. We thus successfully created DNA origami-based 2D assembly systems. The method can be extended to the construction of any structure by programmed self-assembly.
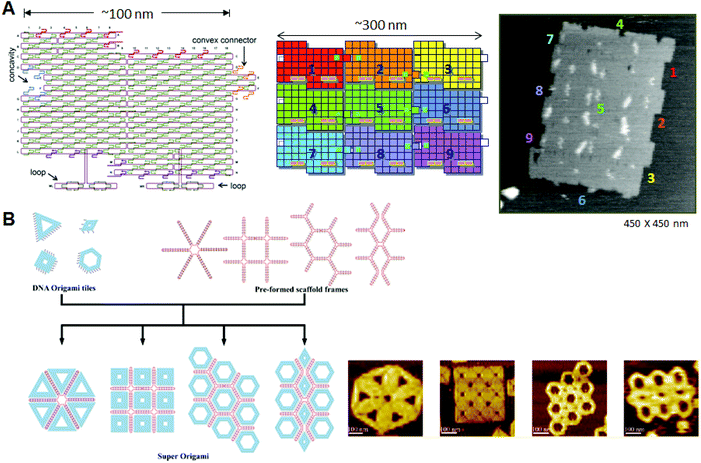 | ||
| Fig. 2 Programmed self-assembly of DNA origami. (A) Structure of DNA origami having a concavity and a convex connector; the structure is called a “DNA jigsaw piece” for 2D assembly. A 3 × 3 assembly of nine origami tiles and the AFM image of the assembly. (B) Programmed assembly of multiple DNA origami structures using the assistance of scaffold frames. Target assemblies and their AFM images are shown. | ||
Recently, Rothemund and co-worker created a programmed assembly system by controlling the positions of adhesive π-stacking terminals for selective connection between rectangular tiles.13 They showed that a relaxed edge with blunt ends can form a stable connection, as opposed to a stressed edge with the usual loop ends, which induce structural distortions. Multiple two-dsDNA terminals with blunt ends were introduced to assemble complementary edges of the counterpart tiles as a binary code. In addition, the complementarity of the edge shape worked effectively to precisely align the different tiles for the one-dimensional assemblies. The results indicate that the π-stacking interactions between the complementary edges can control the programmed assembly of multiple different origami tiles.
Yan and co-workers presented template-assisted assembly of DNA origami structures.14 In this method, scaffold frames prepared from single-stranded template DNA and staple strands were used to direct the positioning of 6–10 predesigned DNA origami structures, including triangles, squares, and hexagons (Fig. 2B). By annealing the origami structures with connection strands and a scaffold frame, the target assemblies were obtained in a predesigned fashion in relatively high yields.
The above strategy can be used to produce a larger assembly when applied to an origami assembly, and the positioning of the origami units can be programmed using the defined design of the origami structures. The variety of available 2D origami structures was expanded by the introduction of the predesigned and template-assisted strategies.
4. 3D DNA origami structures
Given the geometry of the periodic double-helical DNA structure, 3D structures can be designed by extending the 2D DNA origami system. Two strategies for preparing 3D DNA origami structures have been developed: one is the bundling of dsDNAs, where the relative positioning of adjacent dsDNAs is controlled by crossovers, and the other is the folding of 2D origami domains into 3D structures using interconnection strands. In the former method, developed by Shih and co-workers, relative positioning of adjacent dsDNAs is geometrically controlled by the crossovers. By arranging the positions of the crossovers, tubular and multilayered structures can be constructed (Fig. 3A).15 By increasing or decreasing the number of base pairs between crossovers (in this case, two helical rotations for 21 base pairs), the relative positional relationship between adjacent dsDNAs is controlled. Using the rotational angle of 240° for seven base pairs, three adjacent dsDNAs can be placed at a relative angle of ±120° with crossovers every 7 or 14 base pairs. By alternating this relative positioning between adjacent dsDNAs, the duplexes form a pleated structure. When adjacent dsDNAs are placed to rotate in one direction, the contiguous duplexes finally form a six-helix bundled tubular structure. Thus, when some parts of the pleated structures are turned backward by the introduction of one-directional rotation of adjacent dsDNAs, the structures fold to become a stacked layer structure. In this case, to stabilize the 3D structures, adjacent layers of dsDNAs should be further connected by crossovers. Due to the complexity and high density of the introduced crossovers, accurate folding into the target 3D structure requires a week-long folding time. When the pleated structures were integrated as multilayered structures, the repeating units of the six-helix bundled tubular structures formed a honeycomb lattice, viewed from the axial direction of the double helices. It was also possible to create more complex structures by perpendicularly joining these 3D structures. In addition, a wireframe icosahedron structure was assembled from three double-triangle monomers made of a six-helix bundled tubular structure with connections. caDNAno software, which is publicly available, has been developed to support the design of these 3D structures.16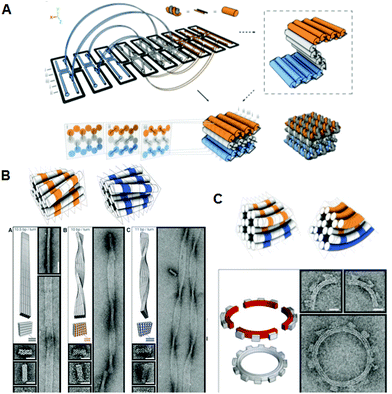 | ||
| Fig. 3 Design and construction of 3D DNA origami structures. (A) Scheme for folding the 2D pleated structure into a 3D multilayered structure using staple strands connecting adjacent layers. Sectional views of the positions of the crossovers in the multilayered structure sliced at seven-base-pair intervals. (B) Global twisted structures of six-helix DNA bundles obtained by the selective deletion or insertion of nucleotides to change the helical turns from the normal 10.5 base pairs to 10 or 11 base pairs. TEM images of the polymerized ribbons containing 10.5-base-pair, 10-base-pair, and 11-base-pair helical pitches. (C) Global bending of six-helix DNA bundles by the deletion and insertion of nucleotides in the adjacent duplexes. Assembly of four components of a quarter circle with three teeth (50 nm radius) and TEM images of the 12-tooth gear. | ||
Furthermore, using the layered structures described above, new 3D structures were built by changing the helical twist from the average helical pitch of 10.5 base pairs per turn to 10 or 11 base pairs per turn.17 When dsDNAs having different helical pitches were bundled together, torque and repulsion between base pairs caused overall structural changes including twisting or 30–180° bending (Fig. 3B). Using these structures as building blocks, left-handed or right-handed helical ribbon structures were prepared. In addition, when angle-controlled duplex bundles were connected to each other, a six-tooth gear and a spherical wireframe capsule were created (Fig. 3C).
Using a different strategy, a DNA box structure was created by folding multiple 2D origami domains with interconnecting strands.18 Six independent rectangles were sequentially linked, and were designed to be folded using interconnection strands in a programmed fashion (Fig. 4A, B). Analyses of the assembled structure by AFM, cryo-electron microscopy (cryo-EM), dynamic light scattering, and small-angle X-ray scattering indicated that the size was close to the original design. The lid of the box could be opened using a specific DNA strand to release the closing duplex by strand displacement, and the opening event was monitored by fluorescence resonance energy transfer (FRET) (Fig. 4C). Other types of DNA boxes have been created using a similar method, which can control both the inside and outside by adjusting the directions of the crossovers at the connection edges.19 A tetrahedral structure was designed and constructed from four aligned origami triangles, which were preconnected with an M13 scaffold strand without folding independent 2D plates.20
 | ||
| Fig. 4 Design and construction of 3D structures from sequentially connected multiple rectangular plates. (A) DNA box structure by folding of six DNA origami rectangles using interconnection strands introduced at the edges of rectangles. (B) The DNA box model reconstructed from cryo-EM images. (C) Controlled opening of the box lid using selective DNA strands (key). Lid opening event was monitored by FRET. | ||
Using the strategy of folding 2D origami structures, we designed and prepared new hollow prism structures (Fig. 5A).21 To make these structures, we first prepared new DNA origami structures having three, four, or six arms, and then introduced connection strands to these multiarm DNA origami structures to allow them to be folded into 3D structures. After introducing the interconnection strands on the side edges of the arms and annealing, no original 2D structures were observed, and linear, fiber-like structures appeared. The 3D structures constructed using this method were closed tubular and were characterized by AFM. Cryo-EM images revealed that the 3D structures were hollow prisms. Successive AFM scanning of the sample forced the closed structures to open into 2D structures (Fig. 5B). The tube-opening process was observed by high-speed AFM imaging, which could successively obtain one AFM image per second. The prism structures completely opened, with the number of scans necessary ranging from 1 to 30 depending on the individual prism. The opened prisms never reverted to their original multiarm structures. After opening, the 2D structure formed a single rectangle of ca. 130 nm × 90 nm, and the scaffold strand could be observed on the opened 2D structure.
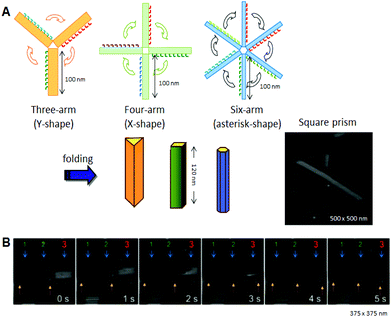 | ||
| Fig. 5 Design and construction of prism structures and observation of their opening events. (A) Construction of various hollow prism structures by folding of multiple rectangular arms with connection strands. The AFM image of the square prism structures. (B) The opening event of a prism (no. 3) visualized by successive high-speed AFM scanning (1 image per second). | ||
We further designed and constructed a cuboid structure using the design of a square prism structure.22 We used the DNA origami method to prepare six rectangular DNA origami plates which were folded into 3D box structures using interconnection strands. Six rectangular plates were designed, and the connection strands were introduced at the sides of the top and bottom plates to enable the prism structure to be closed into a box structure with a size of 36 nm × 36 nm × 44 nm. The six rectangles were formed after annealing without connection strands. After annealing the staple strands with connection strands, the closed structures were observed by AFM. Dynamic light scattering analysis showed the structure to be monodisperse (89%) and about 68 nm in diameter, which was similar to the designed size (67 nm). The box-opening event was observed using high-speed AFM. Successive scanning of the sample converted the closed 3D structure into 2D structures. The morphological changes occurred over 10 s, which was a detectable time scale for our AFM instrument. AFM also enabled us to identify the individual plates involved in the opening process. The time scale for morphological changes of the 3D structures was slow enough to be observed and characterized by high-speed AFM imaging. Dimensional conversion of two dimensions into three dimensions could become a foundation technology for the manipulation of nanostructures. It is expected that target molecules will be caged in and released from the designed nanospace, and that chemical and enzymatic reactions inside the 3D structures will be realized.
5. Modification and functionalization of 2D DNA origami structures
5.1 Selective placement of functional groups
One of the most important features of DNA origami is that each individual position on the 2D structure contains different sequence information. This means that the functional molecules and particles that are attached to the staple strands can be placed at desired positions on the 2D structure.By conjugating ligands and aptamers to staple strands, proteins were selectively attached on the 2D structures.23–26 Combinations of specific proteins and ligands, such as SNAP-tag and halo-tag, were also used for the selective placement of fusion proteins on the DNA origami.27
Yan and co-workers incorporated single-stranded DNAs to detect target RNA molecules on the DNA origami surface at the single-molecule level by AFM (Fig. 6).28 Even though samples containing large amounts of cell-derived RNA were used, only the binding of the target RNA could be visualized, and nonspecific binding was not observed. Once DNA origami tiles carrying different types of complementary DNAs were labeled with the corresponding hairpin DNA markers, binding of RNA targets could be identified from the specific hairpin markers on the DNA origami, even though the different origami tiles were mixed. In this study, the detection limit of RNA molecules was about 1000 molecules, meaning that the target RNA could be directly detected from a single cell without using polymerase chain reaction (PCR) amplification.
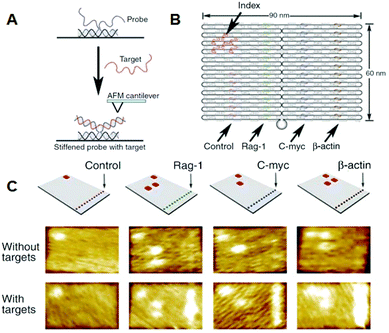 | ||
| Fig. 6 Detection of target RNA by hybridization with probe DNA strands introduced on the DNA origami. (A) The method for imaging the hybridization of target RNA to a probe DNA on the DNA origami. (B) Multiple DNA probes complementary to the target RNAs were introduced onto the DNA origami and hairpin DNAs were also introduced as an index for identifying the probe strand. (C) AFM images of binding of target RNA to the probe strands. Specific DNA probes can be identified by the corresponding index. | ||
Recently, functional molecules have been directly placed using sequence-selective ligands. Pyrrole-imidazole polyamide is a sequence-specific DNA recognition molecule, which binds to the target sequence in the minor groove according to a recognition rule.29 We used a DNA origami structure with five cavities, into which five different sequences were incorporated.30 The alkylation in the anchoring of the polyamide to the selective sequence and then the combined structure were visualized by streptavidin labeling using the conjugating polyamide with biotin. Employing this method, polyamide was found to alkylate the target sequence with 88% yield by discriminating a one-base mismatch. The selective alkylation and subsequent streptavidin labeling revealed the sequence selectivity of the polyamide at the single-molecule level.
Zn-finger protein is another sequence-selective molecule, and the binding sequence can be selected by designing the amino acid sequences.31,32 Using DNA origami containing five cavities, we introduced the strands with recognition sequences into each cavity.33 The Zn-finger proteins bound directly to the cavity containing the target sequence with 50–80% yield. In addition, green-fluorescence-protein-fused Zn-finger proteins were examined for binding to the target sequences. The fusion of the protein lowered the yield of the binding, however, the recognition ability was maintained. These results indicate that Zn-finger proteins can be used as a carrier for positioning the target proteins.
These studies show that the designed sequence-selective recognition molecules can be used to place the functional molecules and proteins on addressable DNA origami structures.
5.2 Single-molecule chemical reactions
Selective bond cleavage and bond formation reactions were performed on a DNA origami surface. Target organic molecules with specific reactivity were introduced into specific positions on the DNA origami. Reductive cleavage of disulfide bonds and oxidative cleavage of olefin by singlet oxygen were carried out on the DNA origami surface, and the reactions proceeded quantitatively at the single-molecule level.34 In addition, amide bond formation and click reactions were performed with 80–90% yield, and three successive reactions were also performed (Fig. 7). These chemical reactions were monitored by the cleavage of biotin-attached chemical linkers and bond formation with biotin-tethered functional groups, which can be labeled with streptavidin for visualization by AFM.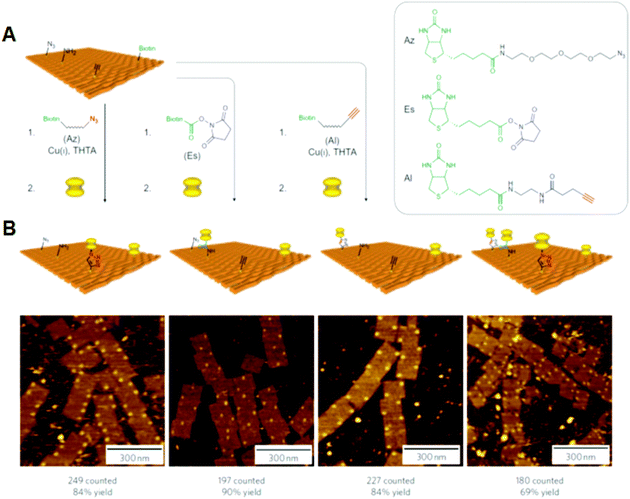 | ||
| Fig. 7 Single chemical reaction on DNA origami. (A) Reactive groups (azido, amino, and alkyne groups) were incorporated into the DNA origami by conjugation with staple DNA strands. The coupling reactions were then performed using the biotin-attached functional groups. The completion of the reactions was visualized by the binding of streptavidin. (B) AFM images of the three individual reactions and three successive reactions by the treatment of three biotin-attached functional groups. Yields are presented below. | ||
5.3 Selective incorporation of nanomaterials
The position-controlled placement of nanomaterials has been carried out by employing the addressable property of DNA origami. By directly coupling gold nanoparticles (AuNPs) with a staple DNA strand, AuNPs have been selectively placed on DNA origami.35,36 Alternatively, thiol groups were placed on DNA origami structures to assemble AuNPs at predesigned positions.37,38 The yield of the binding of AuNPs improved when the small cavities of the origami structures were used to accommodate them. Incorporated single-stranded DNAs on DNA origami controlled the positioning of two DNA-modified carbon nanotubes into cross-junctions on both sides of the DNA origami.39 The position-controlled carbon nanotubes were applied in order to create a single-molecule device, which showed field-effect transistor-like behavior.5.4 Placement of DNA origami onto a fabricated solid surface
In another development, DNA origami technology met top-down nanotechnology, including semiconductor processing. Fine triangular origami binding sites on the surface were fabricated by electron beam lithography and dry etching. Origami binding sites on a SiO2 surface passivated with single-layer trimethylsilane (TMS), or origami binding sites on diamond-like carbon (DLC), were used for the alignment of triangular origami tiles. For the addition of triangular origami, origami tiles were selectively aligned to the binding sites depending on the size of the sites (Fig. 8).40 The hydrophilicity of the surface of the binding sites passivated with TMS is considered to be a driving force for binding the DNA origami, whereas the interactions with binding sites on the DLC remained unclear. Using the various shapes of binding sites on the surface, multiple origami triangles were attached selectively depending on the shape of the binding sites. Using a similar method, gold particles bound to the three vertices of a DNA triangle were successfully aligned to the origami binding sites with controlled orientation and arrangements.40,41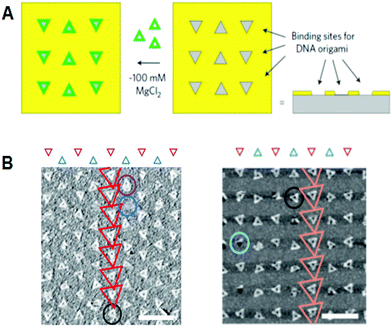 | ||
| Fig. 8 Alignment of DNA triangles onto the fabricated surfaces. (A) Fabricated triangular origami-binding sites were used for the alignment of DNA triangles. (B) Binding of DNA triangles onto the binding sites of TMS-layered SiO2 surface (left) and DLC surface (right). Scale bar 500 nm. | ||
Using the techniques developed in these studies, functional molecules and nanoparticles can be selectively placed at specific positions on the DNA origami in a programmed fashion. The DNA origami system can be integrated with top-down nanotechnology. These methods can be applied to create nanoscale devices with novel functionality, when the functional origami is precisely integrated on the fabricated surface.
6. Application to single biomolecule imaging
6.1 Control of DNA methylation and DNA repair in the DNA nanospace
Direct observation of enzymes interacting with DNA is expected to be one of the most significant technologies for investigating the mechanical behavior of enzymes. Because DNA origami is used as a scaffold for AFM observation, the movement of biomolecules, including proteins and enzymes, and the DNA structural change itself can be visualized and analyzed if the substrate dsDNAs and the target DNA structures are attached onto the origami scaffold.DNA modification using enzymes often requires bending specific DNA strands to facilitate the reaction. The DNA methylation enzyme EcoRI methyltransferase (M.EcoRI) bends dsDNA by 55–59° during the methyl-transfer reaction.42 To control the methyl-transfer reaction of M.EcoRI and examine the structural effect on methylation, we designed and prepared a 2D DNA scaffold, named a “DNA frame”, which accommodates two different lengths of dsDNA fragments: a tense 64-mer dsDNA and a relaxed 74-mer dsDNA (Fig. 9).43 High-speed AFM revealed the different dynamic movements of the dsDNAs and complexes of M.EcoRI with 64-mer and 74-mer dsDNAs. After treatment of the dsDNA in the DNA frame with M.EcoRI and subsequent digestion by the restriction enzyme EcoRI, AFM analysis revealed that, compared with the 64-mer dsDNA, the 74-mer dsDNA was less effectively cleaved, indicating that the methylation preferentially occurred in the relaxed 74-mer dsDNA, rather than in the tense 64-mer dsDNA. Biochemical analysis of the methylation and specific digestion using real-time PCR supported the above results. These results indicate the importance of structural flexibility in the bending of dsDNA during the methyl-transfer reaction with M.EcoRI. Therefore, DNA methylation can be regulated using the tension-controlled dsDNAs incorporated in the DNA frame nanostructure.
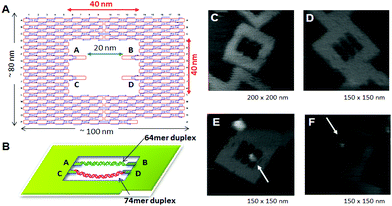 | ||
| Fig. 9 Control of the enzyme reactions in the DNA origami scaffold. (A), (B) DNA frame structure designed for incorporation of two different dsDNAs; tense 64-mer dsDNA and relaxed 74-mer dsDNA having the specific sequence for M. EcoRI at the center. AFM images of DNA frame (C), two-dsDNA attached DNA frame (D), and M. EcoRI bound to the 64-mer dsDNA (E) and the 74-mer (F) dsDNA. | ||
We next developed a novel method for the analysis of DNA repair by employing a DNA frame containing various dsDNAs and using high-speed AFM.38 We employed DNA base excision repair enzymes, 8-oxoguanine glycosylase44 and T4 pyrimidine dimer glycosylase,45 for the analysis of a reaction on the defined DNA nanostructure. These enzymes have glycosylase/AP lyase activity for removing damaged nucleobases and cleaving the DNA strand.46 We placed various dsDNAs with a damaged base onto a DNA nanochip as a dsDNA cassette and analyzed the repair reaction at the single-molecule level. We placed two different lengths of substrate dsDNAs, tense 64-mer and relaxed 74-mer dsDNAs, onto a DNA frame to examine the structural effect on the glycosylase/AP lyase activity, including cleavage of the DNA strand and trapping of reaction intermediates. The relaxed 74-mer dsDNA trapped the enzymes with NaBH4 reduction and was cleaved more effectively compared with the 64-mer dsDNA. In addition, dynamic movement of the enzymes and the single DNA repair reaction were directly observed in the DNA frame using a high-speed AFM. The DNA frame system serves to elucidate the detailed properties of the repair enzymes by direct observation of the events involved in DNA repair.
This method can be used for other DNA-modifying and repair enzymes that bend the double helix during the enzymatic reaction. The method can be extended to the direct observation of various enzymatic phenomena in the designed nanoscale space.
6.2 Visualization of DNA structural changes
The formation and disruption of a single G-quadruplex structure were observed in a nanospace.47 In that study, we employed a DNA frame scaffold. To place the G-rich sequences, we prepared two unique DNA strands (G-strands) which contained single-stranded G-rich overhangs in the middle for the formation of an interstrand G-quadruplex (Fig. 10). Three G-tracts were placed in the upper G-strand whereas the lower strand had a single G-tract.48 The introduced strands were not in contact with each other. In the presence of K+, the G-strands in the DNA frame clearly had an X-shaped structure, which indicated the formation of an interstrand G-quadruplex (Fig. 10, right panel). The efficiency of the X-shaped formation was 44% using relaxed dsDNA. We further directly observed the dynamic formation of the G-quadruplex in real time by high-speed AFM. During scanning of the sample in the presence of K+, the two G-strands maintained a separated state for a given period and then they suddenly formed an X shape. In a similar fashion, we observed the disruption of G-quadruplexes in the absence of K+. The X shape remained unchanged for a while, and then reverted to the separated state under AFM scanning.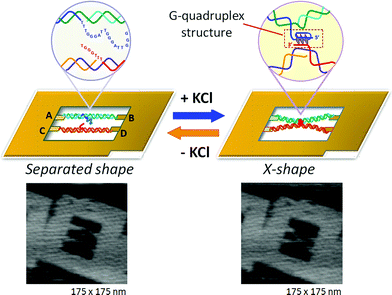 | ||
| Fig. 10 Visualization of G-quadruplex formation using the structural change of two dsDNAs placed in the DNA frame. In the presence of KCl, the separated state changes to the X-shape by connection at the center of two dsDNAs via G-quadruplex formation. | ||
We thus realized the dynamic formation and disruption of G-quadruplexes by monitoring the structural changes of two dsDNAs in the DNA nanostructure through high-speed AFM imaging. This was the first report of the real-time observation of a reversible conformational change of a DNA structure. The method can be applied to the direct observation of various known conformational changes in nucleic acids.
7. Application to DNA molecular machines
One of the goals of the artificial molecular system is fully controllable movement of molecular machines. DNA molecular machines are operated by adding and removing specific DNA strands for complex movements. For this purpose, an extra sequence called a “toehold” is attached to the end of the DNA strand. When a DNA strand fully complementary to a toehold-containing strand is added, the initial strand containing the toehold is selectively removed at the toehold by strand displacement. The thermodynamic stabilization energy works as “fuel” during hybridization to provide the mechanical motion of DNA machines. Using this strategy, DNA tweezers that perform close–open motions were constructed.49 Also created were two examples of a DNA walking device: a DNA walker with two legs that can control its direction of motion and a DNA motor that can move forward autonomously by the cleavage of a DNA-nicking enzyme.50Seeman and co-workers developed a controllable mechanical rotation device called a “PX–JX2 device”, which can rotate two adjacent ends of dsDNAs by 180° in either the PX (paranemic crossover) or JX2 (its topoisomer) state.51–53 Two PX–JX2 devices were introduced onto the origami scaffold at two specific sites to capture DNA nanostructures with four sticky ends through the two sets of two complementary strands on the PX–JX2 devices.54 The PX and JX2 states of the two devices allowed the binding of four different capture molecules with four different patterns of sticky ends corresponding to each rotational operation. Large DNA triangles were introduced onto the capture molecules as AFM markers for identification. Through the use of the four combinations of the PX and JX2 states, the specific capture molecules can be trapped between the devices. In addition, one target capture molecule in a mixture of four different capture DNAs can correctly bind to the specific state of the device after heating to dissociate nonspecific capture molecules (error correction).
Seeman and co-workers also examined the movement of a two-legged DNA walker along tracks constructed in DNA nanostructures55 and created a new DNA walker to operate the assembly line on DNA origami (Fig. 11A).56 Three PX–JX2 devices were fixed onto DNA origami, and the DNA walker moved along a pathway from the outside by following a predetermined operation. The movements of all the devices and the DNA walker were fully controlled by specific DNA strands. PX–JX2 devices carrying AuNPs of various sizes and amounts could pass AuNPs to the walker by a rotational operation when the DNA walker was located nearby. The DNA walker moved in one direction along the track. Three PX–JX2 devices delivered AuNPs to the walker, and the DNA walker then picked up the AuNPs at specific positions. The DNA walker, which eventually picked up three AuNP-bound DNAs, had a 43% yield. As the on–off operation of the PX–JX2 device delivering AuNPs was fully controlled by the specific DNA strand, the final target product was obtained in a high yield (90%) with a very low error rate (1%). Moreover, using the on–off operation of three PX–JX2 devices, eight patterns of AuNPs bound to the DNA walker were obtained.
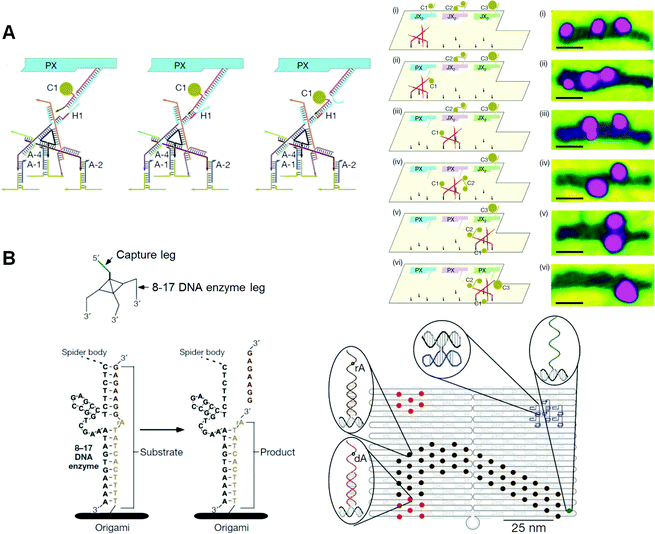 | ||
| Fig. 11 DNA nanomachines walking on the DNA origami. (A) Assembly line of gold nanoparticle (AuNP) using a DNA walker. The DNA walker has four feet for walking on the selective position on the DNA origami and three hands to capture AuNPs. AuNP is transferred to the DNA walker from the PX–JX2 device in the PX state by toehold-hybridization. The DNA walker stops at three specific places to capture AuNPs in PX state. Stepwise AFM images of AuNPs attached on the DNA walker after the operations. (B) DNA spider has three legs consisting of a DNAzyme to cleave the RNA (rA)-containing DNA strand introduced on the DNA origami. The DNA spider starts by releasing from the starting point, which is placed with the capture leg, and then walking on the substrate track by cleaving the substrates. The walker stops when it encounters the uncleavable substrate strands lacking RNA (dA). | ||
Stojanovic, Yan, and co-workers created a DNA nanomachine called a “DNA spider”, which has three legs and one capture strand and moves along various patterned tracks constructed on DNA origami (Fig. 11B).57 A DNAzyme that can hydrolyze RNA was incorporated into the three legs of the DNA spider. Single-stranded DNA/RNA chimeras were incorporated into the DNA origami as a track. A DNA spider was introduced and fixed at the starting position on the DNA origami via a capture strand, and was then released from the starting point using a specific DNA strand. The DNA spider was then bound to the DNA/RNA chimeric strands in the track, and migrated along the predetermined track by cleaving DNA/RNA chimeric strands using DNAzymes in the legs. Finally, the spider stopped at a specific DNA strand, which did not contain cleavable RNA. This study shows that start, walk, and stop operations can be programmed into the predesigned track and DNA spider. The movements of the DNA spider were analyzed in real time by high-resolution total-internal-reflection microscopy, and the spiders were found to have moved across the DNA origami at 3 nm min−1.
We have created a DNA transportation system in which a DNA nanomachine can move along a designed track constructed in DNA origami (Fig. 12). A track on a DNA scaffold was constructed for observation of the multistep movement of a specific DNA strand.58 Multiple ssDNAs (stators) were introduced as a track for hybridization of a complementary strand (motor strand). When the motor strand hybridizes to the specific stator, subsequent cleavage of the stator/motor duplex by a nicking enzyme Nt.BbvCI removes the short ssDNA of the stator, and the motor strand binds to the neighboring intact stator by branch migration and then finally steps forward.59 We expected that multistep movement would be achieved by the turnover of the enzymatic reactions. Seventeen stators were introduced onto the DNA origami scaffold as a motor track to observe the movements of the motor strand. The stator/motor duplex was introduced at site 1 of the motor track using a DNA scaffold lacking the staple strand at site 1. After annealing, a single spot was observed at site 1, and the incorporation proceeded quantitatively. The DNA scaffold carrying the stator/motor at site 1 was then incubated for 0–3 h with Nt.BbvCI to examine migration of the motor strand along the DNA motor track. The motor strand was imaged as a single spot of duplex on the DNA origami scaffold. We observed one-directional and time-dependent movement of the motor strand along the motor track. Furthermore, the movement of the motor strand along the motor track was directly observed by high-speed AFM. The stator/motor duplex spot moved forward along the motor track in each AFM image with scanning every 5 s. In kymograph analysis, the distance of the motor-strand movement corresponded to the distance between adjacent stators, indicating that the movement occurred stepwise on the track.
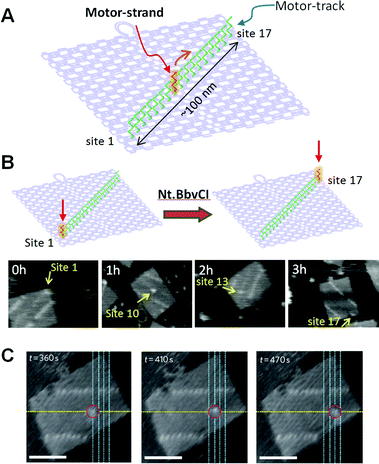 | ||
| Fig. 12 DNA motor moving on the DNA origami. (A) A motor-track (green ssDNAs) was constructed on the DNA origami and the movement of the DNA motor (red ssDNA) was examined. (B) Time-dependent movement of a DNA motor. (C) Stepwise movement of a DNA motor observed in real-time by high-speed AFM. Scale bar 50 nm. | ||
We further designed a complicated system for the control of the direction of the motor movement using a branched track and controllable blocking strands.60 A branched track was constructed on DNA origami, and three branching points and four final destinations were created. Blocking strands were introduced on both sides of the branching points to control the direction of the DNA motor. When one side of the blocking strand was removed by the corresponding release strand, the motor strand could pass the branching point in the predefined direction. The DNA passed two branching points, therefore the two releasing strands determined the pathway and destinations in a programmed fashion. The arrival at final destinations was observed by AFM and fluorescence quenching. The DNA motor found the predefined destinations by following the programmed instructions.
We thus designed and constructed a controllable DNA motor system using a track on a DNA origami scaffold. The stepwise movement of the motor strand was directly observed by high-speed AFM imaging. In addition, the direction of the movement was controlled by blocking and releasing of the specific strands. This method can be applied to the development of a nanomachine that can transport specific molecules and perform more complicated movements on expanded DNA nanostructures.
8. DNA nanostructures for cellar applications
The various applications of DNA origami nanostructures described above have great biological potential and have already been extended to cellular studies. A few examples of DNA nanostructures being resistant to various types of endo- and exonucleases have been reported.61 DNA origami constructs were able to maintain their integrity without degradation or damage in cell lysate of a series of cell lines.62 The high stability of DNA nanostructures in a biological system and their favorable compatibility with functional biomolecules, such as proteins and aptamers, demonstrate that the nanostructures are promising biomaterials for living-cell analysis and as platforms for safe drug delivery. Unmethylated cytosine-phosphate-guanine (CpG) oligonucleotides with strong immunostimulatory activities can be recognized by endosomal Toll-like receptor 9 (TLR9) to induce immunostimulatory responses of the immune cells.63,64 Multiple-branched DNA nanostructures and DNA tetrahedra bearing CpG motifs were developed for noninvasive intracellular deliveries.65,66 Furthermore, taking advantage of a similar triggering mechanism, Liedl and co-workers designed and constructed hollow tube-shaped origami which served as an active carrier system for the investigation of immune responses in mammalian cells.67 The designs of the origami structure and the endocytotic pathway are illustrated in Fig. 13. One hollow DNA origami structure (∼80 nm × 20 nm) with maximized surface area had as many as 62 inner or 62 outer binding sites (handle sequences, Hs) for CpG and anchor sequences (CpG Hs). The delivery performances and immunostimulatory responses of the DNA tubes holding CpG were investigated in freshly isolated mouse splenocyte cells. The origami tube was first taken up by immune cells, and then fused with a vesicle containing TLR9 segregated by a Golgi apparatus. The origami tube with CpG sequences was recognized by TLR9 in a vesicle, inducing the immune signaling cascade. Finally, a series of molecules, such as cytokines and CD69, were expressed to further stimulate the immune response.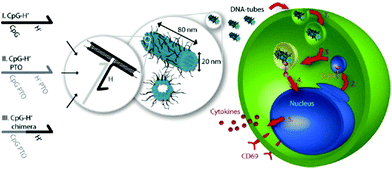 | ||
| Fig. 13 Schematic design of DNA origami tube and endocytotic pathway. Left: Three different types of CpG-H′ designed to hybridize with CpG. Right: endocytotic pathway of tube origami holding CpG with immune cells and subsequently stimulating the immune responses. | ||
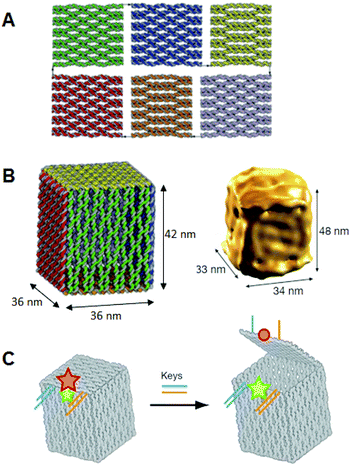
Besides intracellular signal triggering, Douglas and co-workers created a hexagonal barrel called a DNA nanorobot (shown in Fig. 14A) for transporting molecular payloads to target cells and subsequent multiple interactions with the cellular system.68 By employing an aptamer-based locking mechanism, the reconfiguration of the nanorobot structure for payload delivery was triggered using sensing of molecules on the cell surface as signal inputs. Aptamer-complement duplexes were introduced on the left and right sides of the front of the barrel. This hexagonal barrel nanodevice can be unlocked in response to the protein keys between an aptamer-complement duplex and aptamer–target complex (Fig. 14B). The hollow inside can be loaded with different types of payloads, such as proteins and nanoparticles (Fig. 14C, D), in a highly organized fashion for the delivery of various payloads, and the innate conformational regulation of the robot can be controlled selectively. To obtain a high yield of the nanorobot in a closed state, two “guide” staples were incorporated close to the locking site (Fig. 14E). The aptamer locking mechanism was designed to play the role of a logic gate by binding to the target cells. When the same aptamer sequences are used at two locking sites, the robot can be activated in response to only one type of key, whereas two types of inputs (cell surface antigens) as keys are required at the same time if two different aptamer sequences are employed in order to activate the robot's function and expose the payload for further interactions with target cells (Fig. 14F). Furthermore, this robot can be used to interface with cells and stimulate their signaling in an inhibition or activation manner by selective regulation of the nanorobot function. Therefore, DNA origami affords a new strategy for applications in cellular studies.
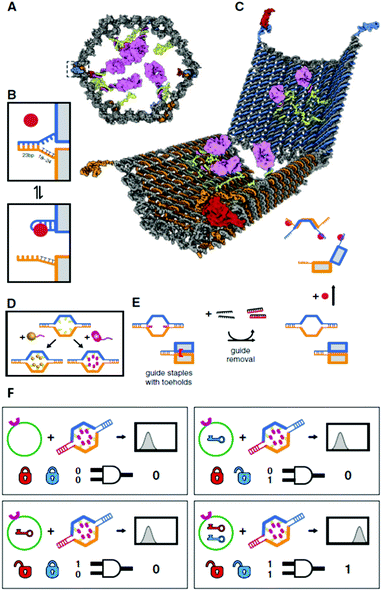 | ||
| Fig. 14 Design of aptamer-gated DNA nanorobot. (A) Schematic drawings of the closed nanorobot in hexagonal barrel shape loaded with protein payloads inside in front orthographic view. The nanodevice was fastened by two DNA aptamer based locks on the left (dashed box) and right side. (B) The nanodevice is unlocked when a DNA aptamer (orange) and the complementary strand (orange) dissociate in the existence of antigen key (red). (C) Schematic view of the open state of nanorobot by protein key displacement of aptamer locks. (D) Gold nanoparticles (gold) and Fab′ antibody fragments are employed as payloads inside of the nanorobot after modifications. (E) Guide staples with toeholds are incorporated to obtain the high yield of nanorobot in the closed state. (F) AND-gated nanorobot is activated (aptamer-encoded unlock) by molecular inputs expressed by target cells. | ||
9. Conclusion and prospects
A wide variety of designs of 2D and 3D structures of around 100 nm in size have been realized by constructing DNA origami. Compared with the use of small DNA assemblies, the DNA origami method reduces experimental labor and the need for strict stoichiometry and eliminates uncertainties. In the relatively short time since the DNA origami method was first reported in 2006, multidimensional structures, functionalization, single-molecule imaging, and the construction of molecular machines have been realized. Functionalized DNA origami has already been combined with top-down nanotechnology, including semiconductor processing techniques. As the size of DNA origami is compatible with cellular uptake, it is expected that cell-targeting applications will benefit from the design of various shapes and effective functionalizations.It would be possible to use functional DNA origami as a module to express higher-level functionalities by assembling them in a programmed fashion. It is extremely difficult to arrange small molecules in order to prepare desired structures. However, if preassembled 100 nm-sized structures can be organized and purified to exclude incomplete structures, the usefulness of the structures will greatly improve.
DNA origami allows the precise placement and manipulation of functional molecules and biomolecules and is available for the construction of a designed nanospace for chemical and biological reactions. Furthermore, as the observation of the movement of biomolecules in a nanospace is now possible, it is also possible to create devices that visualize the reaction and the behavior of biomolecules in the designed nanospace. This technology also opens the way to expressing the complex functionality of the programmed organization of many different modules seen in living systems.
Notes and references
- N. C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS.
- N. C. Seeman, Nature, 2003, 421, 427–431 CrossRef.
- M. Endo and H. Sugiyama, Chem.–Eur. J. Chem. Biol., 2009, 10, 2420–2443 Search PubMed.
- A. Rajendran, M. Endo and H. Sugiyama, Angew. Chem., Int. Ed., 2012, 51, 874–890 CrossRef CAS.
- T. Torring, N. V. Voigt, J. Nangreave, H. Yan and K. V. Gothelf, Chem. Soc. Rev., 2011, 40, 5636–5646 RSC.
- P. W. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS.
- B. Hogberg, T. Liedl and W. M. Shih, J. Am. Chem. Soc., 2009, 131, 9154–9155 CrossRef CAS.
- E. Pound, J. R. Ashton, H. A. Becerril and A. T. Woolley, Nano Lett., 2009, 9, 4302–4305 CrossRef CAS.
- Z. Zhao, H. Yan and Y. Liu, Angew. Chem., Int. Ed., 2010, 49, 1414–1417 CAS.
- M. Endo, T. Sugita, Y. Katsuda, K. Hidaka and H. Sugiyama, Chem.–Eur. J., 2010, 16, 5228 CrossRef.
- A. Rajendran, M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, ACS Nano, 2011, 5, 665–671 CrossRef CAS.
- M. Endo, T. Sugita, A. Rajendran, Y. Katsuda, T. Emura, K. Hidaka and H. Sugiyama, Chem. Commun., 2011, 47, 3213–3215 RSC.
- S. Woo and P. W. Rothemund, Nat. Chem., 2011, 3, 620–627 CrossRef CAS.
- Z. Zhao, Y. Liu and H. Yan, Nano Lett., 2011, 11, 2997–3002 CrossRef CAS.
- S. M. Douglas, H. Dietz, T. Liedl, B. Hogberg, F. Graf and W. M. Shih, Nature, 2009, 459, 414–418 CrossRef CAS.
- S. M. Douglas, A. H. Marblestone, S. Teerapittayanon, A. Vazquez, G. M. Church and W. M. Shih, Nucleic Acids Res., 2009, 37, 5001–5006 CrossRef CAS.
- H. Dietz, S. M. Douglas and W. M. Shih, Science, 2009, 325, 725–730 CrossRef CAS.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, M. M. Golas, B. Sander, H. Stark, C. L. Oliveira, J. S. Pedersen, V. Birkedal, F. Besenbacher, K. V. Gothelf and J. Kjems, Nature, 2009, 459, 73–76 CrossRef CAS.
- A. Kuzuya and M. Komiyama, Chem. Commun., 2009, 4182–4184 RSC.
- Y. Ke, J. Sharma, M. Liu, K. Jahn, Y. Liu and H. Yan, Nano Lett., 2009, 9, 2445–2447 CrossRef CAS.
- M. Endo, K. Hidaka, T. Kato, K. Namba and H. Sugiyama, J. Am. Chem. Soc., 2009, 131, 15570–15571 CrossRef CAS.
- M. Endo, K. Hidaka and H. Sugiyama, Org. Biomol. Chem., 2011, 9, 2075–2077 RSC.
- R. Chhabra, J. Sharma, Y. Ke, Y. Liu, S. Rinker, S. Lindsay and H. Yan, J. Am. Chem. Soc., 2007, 129, 10304–10305 CrossRef CAS.
- W. Shen, H. Zhong, D. Neff and M. L. Norton, J. Am. Chem. Soc., 2009, 131, 6660–6661 CrossRef CAS.
- S. Rinker, Y. Ke, Y. Liu, R. Chhabra and H. Yan, Nat. Nanotechnol., 2008, 3, 418–422 CrossRef CAS.
- A. Kuzuya, M. Kimura, K. Numajiri, N. Koshi, T. Ohnishi, F. Okada and M. Komiyama, ChemBioChem, 2009, 10, 1811–1815 CrossRef CAS.
- B. Sacca, R. Meyer, M. Erkelenz, K. Kiko, A. Arndt, H. Schroeder, K. S. Rabe and C. M. Niemeyer, Angew. Chem., Int. Ed., 2010, 49, 9378–9383 CrossRef CAS.
- Y. Ke, S. Lindsay, Y. Chang, Y. Liu and H. Yan, Science, 2008, 319, 180–183 CrossRef CAS.
- T. Bando and H. Sugiyama, Acc. Chem. Res., 2006, 39, 935–944 CrossRef CAS.
- T. Yoshidome, M. Endo, G. Kashiwazaki, K. Hidaka, T. Bando and H. Sugiyama, J. Am. Chem. Soc., 2012, 134, 4654–4660 Search PubMed.
- J. G. Mandell and C. F. Barbas, 3rd, Nucleic Acids Res., 2006, 34, W516–W523 CrossRef CAS.
- J. D. Sander, P. Zaback, J. K. Joung, D. F. Voytas and D. Dobbs, Nucleic Acids Res., 2007, 35, W599–W605 Search PubMed.
- E. Nakata, F. F. Liew, C. Uwatoko, S. Kiyonaka, Y. Mori, Y. Katsuda, M. Endo, H. Sugiyama and T. Morii, Angew. Chem., Int. Ed., 2012, 51, 2421–2424 Search PubMed.
- N. V. Voigt, T. Torring, A. Rotaru, M. F. Jacobsen, J. B. Ravnsbaek, R. Subramani, W. Mamdouh, J. Kjems, A. Mokhir, F. Besenbacher and K. V. Gothelf, Nat. Nanotechnol., 2010, 5, 200–203 CrossRef CAS.
- J. Sharma, R. Chhabra, C. S. Andersen, K. V. Gothelf, H. Yan and Y. Liu, J. Am. Chem. Soc., 2008, 130, 7820–7821 CrossRef CAS.
- B. Ding, Z. Deng, H. Yan, S. Cabrini, R. N. Zuckermann and J. Bokor, J. Am. Chem. Soc., 2010, 132, 3248–3249 CrossRef CAS.
- A. Kuzuya, N. Koshi, M. Kimura, K. Numajiri, T. Yamazaki, T. Ohnishi, F. Okada and M. Komiyama, Small, 2010, 6, 2664–2667 CrossRef CAS.
- M. Endo, Y. Yang, T. Emura, K. Hidaka and H. Sugiyama, Chem. Commun., 2011, 47, 10743–10745 RSC.
- H. T. Maune, S. P. Han, R. D. Barish, M. Bockrath, W. A. Iii, P. W. Rothemund and E. Winfree, Nat. Nanotechnol., 2010, 5, 61–66 CrossRef CAS.
- R. J. Kershner, L. D. Bozano, C. M. Micheel, A. M. Hung, A. R. Fornof, J. N. Cha, C. T. Rettner, M. Bersani, J. Frommer, P. W. Rothemund and G. M. Wallraff, Nat. Nanotechnol., 2009, 4, 557–561 CrossRef CAS.
- A. M. Hung, C. M. Micheel, L. D. Bozano, L. W. Osterbur, G. M. Wallraff and J. N. Cha, Nat. Nanotechnol., 2010, 5, 121–126 CrossRef CAS.
- B. Youngblood and N. O. Reich, J. Biol. Chem., 2006, 281, 26821–26831 Search PubMed.
- M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 1592–1597 CrossRef CAS.
- S. D. Bruner, D. P. Norman and G. L. Verdine, Nature, 2000, 403, 859–866 CrossRef CAS.
- K. Morikawa, O. Matsumoto, M. Tsujimoto, K. Katayanagi, M. Ariyoshi, T. Doi, M. Ikehara, T. Inaoka and E. Ohtsuka, Science, 1992, 256, 523–526 Search PubMed.
- H. M. Nash, S. D. Bruner, O. D. Scharer, T. Kawate, T. A. Addona, E. Spooner, W. S. Lane and G. L. Verdine, Curr. Biol., 1996, 6, 968–980 Search PubMed.
- Y. Sannohe, M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 16311–16313 CrossRef CAS.
- Y. Xu, H. Sato, Y. Sannohe, K. Shinohara and H. Sugiyama, J. Am. Chem. Soc., 2008, 130, 16470–16471 CrossRef CAS.
- B. Yurke, A. J. Turberfield, A. P. Mills, Jr., F. C. Simmel and J. L. Neumann, Nature, 2000, 406, 605–608 CrossRef CAS.
- J. Bath and A. J. Turberfield, Nat. Nanotechnol., 2007, 2, 275–284 CrossRef CAS.
- H. Yan, X. P. Zhang, Z. Y. Shen and N. C. Seeman, Nature, 2002, 415, 62–65 CrossRef CAS.
- S. P. Liao and N. C. Seeman, Science, 2004, 306, 2072–2074 CrossRef CAS.
- B. Ding and N. C. Seeman, Science, 2006, 314, 1583–1585 CrossRef CAS.
- H. Gu, J. Chao, S. J. Xiao and N. C. Seeman, Nat. Nanotechnol., 2009, 4, 245–248 CrossRef CAS.
- T. Omabegho, R. Sha and N. C. Seeman, Science, 2009, 324, 67–71 CrossRef CAS.
- H. Z. Gu, J. Chao, S. J. Xiao and N. C. Seeman, Nature, 2010, 465, 202–U286 CrossRef CAS.
- K. Lund, A. J. Manzo, N. Dabby, N. Michelotti, A. Johnson-Buck, J. Nangreave, S. Taylor, R. Pei, M. N. Stojanovic, N. G. Walter, E. Winfree and H. Yan, Nature, 2010, 465, 206–210 CrossRef CAS.
- S. F. Wickham, M. Endo, Y. Katsuda, K. Hidaka, J. Bath, H. Sugiyama and A. J. Turberfield, Nat. Nanotechnol., 2011, 6, 166–169 CrossRef CAS.
- J. Bath, S. J. Green and A. J. Turberfield, Angew. Chem., Int. Ed., 2005, 44, 4358–4361 CrossRef CAS.
- S. F. Wickham, J. Bath, Y. Katsuda, M. Endo, K. Hidaka, H. Sugiyama and A. J. Turberfield, Nat. Nanotechnol., 2012, 7, 169–173 Search PubMed.
- C. E. Castro, F. Kilchherr, D. N. Kim, E. L. Shiao, T. Wauer, P. Wortmann, M. Bathe and H. Dietz, Nat. Methods, 2011, 8, 221–229 CrossRef CAS.
- Q. Mei, X. Wei, F. Su, Y. Liu, C. Youngbull, R. Johnson, S. Lindsay, H. Yan and D. Meldrum, Nano Lett., 2011, 11, 1477–1482 CrossRef CAS.
- J. Vollmer and A. M. Krieg, Adv. Drug Delivery Rev., 2009, 61, 195–204 CrossRef CAS.
- E. Latz, A. Verma, A. Visintin, M. Gong, C. M. Sirois, D. C. Klein, B. G. Monks, C. J. McKnight, M. S. Lamphier, W. P. Duprex, T. Espevik and D. T. Golenbock, Nat. Immunol., 2007, 8, 772–779 Search PubMed.
- K. Mohri, M. Nishikawa, N. Takahashi, T. Shiomi, N. Matsuoka, K. Ogawa, M. Endo, K. Hidaka, H. Sugiyama, Y. Takahashi and Y. Takakura, ACS Nano, 2012, 6, 5931–5940 Search PubMed.
- J. Li, H. Pei, B. Zhu, L. Liang, M. Wei, Y. He, N. Chen, D. Li, Q. Huang and C. Fan, ACS Nano, 2011, 5, 8783–8789 CrossRef CAS.
- V. J. Schuller, S. Heidegger, N. Sandholzer, P. C. Nickels, N. A. Suhartha, S. Endres, C. Bourquin and T. Liedl, ACS Nano, 2011, 5, 9696–9702 Search PubMed.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831–834 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2013 |
