Non-monotonic cell differentiation pattern on extreme wettability gradients
Marco
Cantini†
*ab,
Maria
Sousa†
acd,
David
Moratal
a,
João F.
Mano
cd and
Manuel
Salmerón-Sánchez
*ae
aCenter for Biomaterials and Tissue Engineering, Universitat Politècnica de València, Valencia, Spain. E-mail: marco1.cantini@mail.polimi.it; masalsan@fis.upv.es
bInstitute for Bioengineering of Catalonia, Barcelona, Spain
c3B Research Group—Biomaterials, Biodegradables and Biomimetics, Headquarters of the European Institute of Excellence on Tissue Engineering and Regenerative Medicine, Department of Polymer Engineering, University of Minho, Guimarães, Portugal
dICVS/3B's—PT Government Associated Laboratory, Braga/Guimarães, Portugal
eCIBER de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Valencia, Spain
First published on 16th October 2012
Abstract
In this study, we propose a methodology to obtain a family of biomimetic substrates with a hierarchical rough topography at the micro and nanoscale that span the entire range of wettability, from the superhydrophobic to the superhydrophilic regime, through an Ar-plasma treatment at increasing durations. Moreover, we employ the same approach to produce a superhydrophobic-to-superhydrophilic surface gradient along centimetre-length scale distances within the same sample. We characterize the biological activity of these surfaces in terms of protein adsorption and cell response, using fibronectin, a major component of the extracellular matrix, and C2C12 cells, a myoblast cell line. Fibronectin conformation, assessed via binding of the monoclonal antibody HFN7.1, exhibits a non-monotonic dependence on surface wettability, with higher activity on hydrophilic substrates (WCA = 38.6 ± 8.1°). On the other hand, the exposition of cell-binding epitopes is diminished on the surfaces with extreme wetting properties, the conformation being particularly altered on the superhydrophobic substrate. The assessment of cell response via the myogenic differentiation process reveals that a gradient surface promotes a different response with respect to cells cultured on discrete uniform samples: even though in both cases the same non-monotonic differentiation pattern is found, the differential response to the various wettabilities is enhanced along the gradient while the overall levels of differentiation are diminished. On a gradient surface cells are in fact exposed to a range of continuously changing stimuli that foster cell migration and detain the differentiation process.
1. Introduction
Surface gradient materials are of foremost interest in regenerative medicine, as they allow one to investigate, on the same substrate, cellular responses to a continuous range of physical, chemical or biochemical stimuli.1 Besides their huge potential as tools for an effective approach to tissue engineering (chemical and/or physical gradients can be directly incorporated into the design of biomaterials to engineer heterogeneous tissues and tissue interfaces), such gradient surfaces are of particular interest for basic studies of the interactions between biological species (proteins, cells) and surfaces, since the effect of a selected property, such as chemical composition,2 stiffness,3 or protein adsorption,4 can be examined in a single experiment on one surface, without introducing experimental artifacts due to discrete substrate preparations.1,5 Among them, the wettability gradient is one of the most interesting and one of the most studied, given its numerous practical applications, such as biomolecular interaction investigations, cell-motility studies, diagnostics, nanotribology, liquid self-transportation, and microfluidics.6–8 Some groups have reported the preparation of wettability gradient surfaces and their uses in studying interaction phenomena with proteins and cells;2,9,10 however, these studies are generally limited to the common hydrophilic–hydrophobic range (i.e., static water contact angle (WCA) changes from about 110° to 20°). There is both fundamental and practical interest in extending such investigations towards extreme wettability properties: superhydrophobicity (WCA greater than 150°) and superhydrophilicity (WCA below 5°);11 these conditions are generally obtained by combining the use of low- or high-surface-energy materials, respectively, with surfaces presenting a hierarchical roughness at both the nano- and microscale.12 Highly water repellent and water adsorbent surfaces could provide new insights on the influence of such extreme environments on the biological response, in terms of protein adsorption and cell behavior. As a matter of fact, the use of this kind of materials has generated an increasing interest in the biomedical community, as indicated by the recent studies addressing cell response on uniform or patterned substrates with extreme wetting properties.11,13–17 The extension of surface wettability gradients to include the superhydrophobic (SH) and the superhydrophilic (sh) limits has been recently explored;11,12,18,19 nevertheless, to the authors’ knowledge, no study has yet addressed cell response along this kind of gradient.Whether it presents a gradient or a uniform surface, a substrate does not interact directly with living cells; this interaction primarily depends on the nature and bioactivity of the layer of extracellular matrix (ECM) proteins that adsorb onto the surface of the substrate upon contact with physiological fluids in vivo or culture medium in vitro.20–23 Cells recognize these proteins via a family of αβ heterodimers, integrins, which provide trans-membrane links between the ECM and the actin cytoskeleton.24 Substrate-bound integrins cluster and develop discrete supramolecular complexes, focal adhesions, that contain important structural proteins (e.g., vinculin, talin, tensin) and signalling molecules (e.g., FAK, Src, paxillin); focal adhesions anchor the cells to the surface and trigger the subsequent cellular response.25
Among the proteins of the ECM that mediate cell adhesion, the importance of fibronectin (FN) was recognized earlier.26 FN is a high molecular weight glycoprotein, consisting of two subunits of 220 kDa, covalently linked by a pair of disulfide bonds near their carboxyl termini; each subunit contains three types of repeating units (termed FN repeats I, II, and III) that mediate interactions with other FN molecules, other ECM proteins, and cell surface receptors. Through these kinds of interactions, FN comes to play a fundamental role in mediating and promoting cell adhesion, and in regulating cell survival and phenotype expression on different substrates.27–34 Indeed, the nature of the surface chemistry has been demonstrated to modulate the amount and the conformation of adsorbed FN.27
In this study, we investigate FN adsorption, in terms of amount and conformation, and the subsequent cell response, through the myogenic differentiation process, along a superhydrophobic-to-superhydrophilic wettability gradient obtained via plasma treatment of a superhydrophobic polystyrene (SH-PS) surface.
2. Results and discussion
2.1 Surface properties
Superhydrophobic polystyrene (SH-PS) surfaces were prepared via a phase-separation methodology according to an established protocol.15,35 Briefly, polymer precipitation onto a smooth PS substrate was forced by immersing a solution of the polymer in a nonsolvent (ethanol), and led to the formation of a rough surface that exhibited high water repellency (Fig. 1a). This superhydrophobic effect is the result of the hierarchical roughness created on the surface of a material with low surface energy (Fig. 2a): spheres with sizes from about 50 to 200 nm agglomerate in larger micrometre structures that prevent the intrusion of the liquid into the valleys of the roughness, in such a way that a water droplet is suspended above the asperities of the surface. This situation is called the Cassie–Baxter state, after the corresponding theoretical model:36,37 the apparent contact angle of the SH-PS results from the contribution of two different phases, one being the solid polymer fraction (PS, WCA = 89.3 ± 1.6°, Fig. 1) and the other the air pockets trapped within the surface roughness (WCA = 180°). Hence, the contact angle measurements of the smooth and rough surfaces suggest that only about 10% of the liquid is in contact with the material surface,17,35 a fact that will surely affect initial protein adsorption and cell behavior at the interface with SH-PS.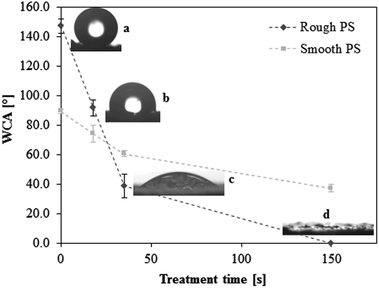 | ||
| Fig. 1 Effect of Ar-plasma treatment on the static water contact angle (WCA) of superhydrophobic rough polystyrene (a, SH-PS, 147.3 ± 4.8°); the chosen treatment times are 18 s (b, SH-PSpl18s, 91.6 ± 5.3°), 35 s (c, SH-PSpl35s, 38.6 ± 8.1°), and 150 s (d, SH-PSpl150s or sh-PS, < 5°). The WCA of smooth polystyrene before and after treatment is also represented (in light grey). The dotted lines are guides for the eye. | ||
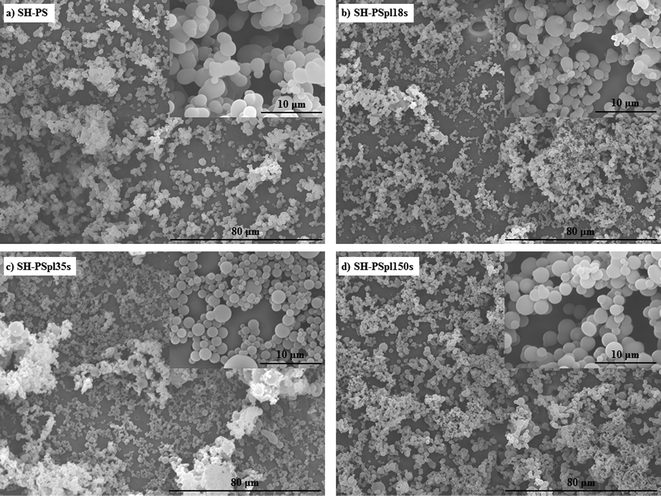 | ||
| Fig. 2 Scanning electron microscopy (SEM) images of SH-PS before (a) and after (b–d) Ar-plasma treatment of different durations, demonstrating that the treatment does not affect the morphology of the samples. | ||
A surface such as the superhydrophobic one that we have generated is prone to chemical modification to form a superhydrophilic substrate; indeed, topographic effects, besides amplifying the intrinsic contact angle of an already hydrophobic surface, can also enhance partial wetting and yield super-wetting properties.38,39 Hence, tuning the surface energy of the substrate by introducing hydrophilic groups, a controlled change of the surface wettability might be obtained. As a method to achieve this goal we used plasma treatment in an argon working atmosphere: a plasma discharge produces radicals on the material surface, forming unstable species that are susceptible to reaction with the oxygen present in the residual air within the plasma chamber, eventually yielding various oxygen-based polar functionalities (hydroxyl, aldehyde and carboxyl groups).11,15,35 The oxidation of the surface allows one to control the wettability by tuning the exposure time to the plasma; having optimized the experimental conditions (base pressure of 18 Pa, argon flow of 100 sccm, power of the plasma discharge of 30 W), we obtained surfaces with progressively decreasing contact angles at increasing treatment times (Fig. 1): starting from the SH surface we obtained a hydrophobic one (WCA = 91.6 ± 5.3°) for treatment time t = 18 s (SH-PSpl18s), a hydrophilic one (WCA = 38.6 ± 8.1°) for t = 35 s (SH-PSpl35s), and finally a superhydrophilic one (WCA < 5°) for t = 150 s (SH-PSpl150s or sh-PS). Moreover, scanning electron microscopy of the substrates revealed that the plasma treatment did not affect the topography of the surface neither at the micro nor at the nano-scale (Fig. 2b–d). Concordantly, the surface roughness, measured via AFM (Rrms = 548 ± 148 nm), was not affected by the Ar-plasma treatment. These results point out the changes in wettability as a sole consequence of the chemical modification induced by the plasma discharge. Indeed, changing the chemical composition of the surface with the introduction of high surface energy groups drives a transition from the Cassie–Baxter state to the Wenzel state,6,12 described as the penetration of the liquid into the asperities of the substrate;40 in this situation, the Wenzel model, which dictates the enhancement of the wetting property of the surface as a linear function of its roughness, applies. In the case of a hydrophilic surface, this implies that the roughness will make it more hydrophilic, eventually determining a superhydrophilic state:37,39 after a 150 s-plasma treatment, the WCA of smooth PS only decreases to 37.1 ± 2.4°, whilst the rough substrate becomes superhydrophilic (Fig. 1). Moreover, the surface roughness becomes impregnated with water as a result of a capillary effect that further improves the wetting (Fig. 1d).38
The methodology applied to produce this family of surfaces that present the same rough topography but controlled wettability, spanning from the SH to the sh limit, can be straightforwardly adapted to the creation of a surface gradient exhibiting this range of wetting properties in one sample. Indeed, using a moving mask that gradually exposes a rectangular SH-PS sample to the plasma discharge, a surface gradient of wettability can be obtained, whose slope is tuned by varying the speed of the mask (Fig. 3a). With a mask moving at 140 μm s−1 a full wettability surface gradient was obtained along a distance of 3 cm (Fig. 3b).
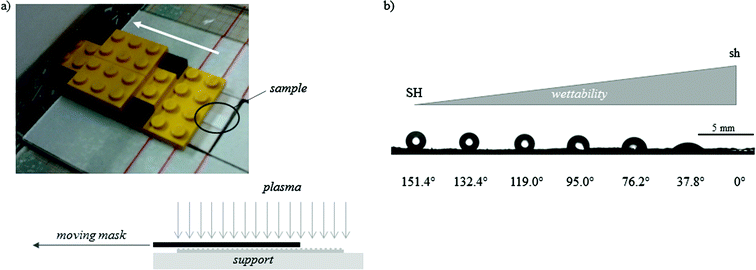 | ||
| Fig. 3 (a) A picture of the moving mask used to obtain a gradual exposition of the sample to the Ar-plasma and a sketch of the system; in the current setting, the mask moves at a constant velocity of 140 μm s−1. (b) A picture of the WCAs along the obtained gradient of wettability (from superhydrophobic, SH, to superhydrophilic, sh), with an indication of their value; the picture is combined from adjacent photographs along the substrate because the angle of view of the CA measurement system is not wide enough. | ||
2.2 Fibronectin adsorption
It is well established that cell adhesion and subsequent response to a synthetic material is mediated by a layer of extracellular matrix (ECM) proteins, which adsorb onto its surface upon contact with physiological fluids in vivo or culture medium in vitro.20,41–43 Hence, fibronectin (FN) adsorption was investigated on the surfaces with different wettabilities produced in this study (SH-PS, SH-PSpl18s, SH-PSpl35s, and sh-PS). The amount of FN adsorbed onto these surfaces, from a 20 μg mL−1 solution, was found to decrease monotonically with increasing wettability (Fig. 4a). FN has been previously reported to adsorb in greater amounts onto hydrophobic rather than hydrophilic surfaces,33,34,44 and our results suggest that this behavior is maintained when extreme wetting properties are included. As a matter of fact, Neto et al. also observed a similar trend of fibronectin adsorption on surfaces ranging from superhydrophobicity to superhydrophilicity.15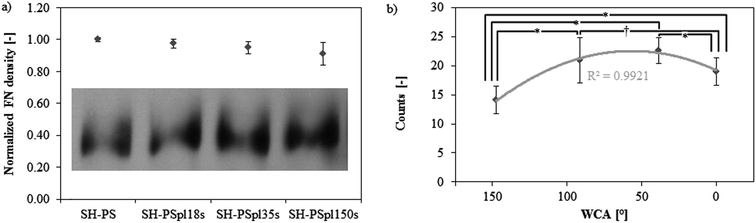 | ||
| Fig. 4 (a) Western blot band of fibronectin remaining in the supernatant after adsorption from a 20 μg mL−1 solution in DPBS and quantification of fibronectin surface density on Ar-treated samples with different treatment times by subtracting the FN in the supernatant from the total protein present in the solution, (FNtot − FNsupernatant)/Asample; the results in the graph are normalized by the value in SH-PS. (b) Monoclonal antibody binding for HFN7.1 measured through ELISA to assess the fibronectin conformation (values are normalized by the surface density of fibronectin). Statistically significant differences between the samples (as determined by ANOVA) are indicated with * (p < 0.05) and † (p < 0.1); the data are fitted by a second-order polynomial regression curve. | ||
With regard to the protein conformation upon adsorption, an enzyme-linked immunosorbent assay (ELISA) with monoclonal antibodies was used to measure the availability of cell adhesion domains, which is a well-established method to probe for structural or conformational changes in adsorbed proteins.45,46 The antibody used, HFN7.1, binds to the flexible linker between the 9th and 10th type III repeats in the central integrin binding domain of FN,28,47 and has been demonstrated to be a receptor-mimetic probe for integrin binding and cell adhesion.27 Results (normalized by the surface density of FN) show a non-monotonic dependence of FN activity on surface wettability: the protein adopts a more favorable conformation on the hydrophilic surface (SH-PSpl35s), whilst the exposure of cell-adhesive epitopes is significantly impaired on the superhydrophobic substrate and is diminished on the hydrophobic (SH-PSpl18s) and on the superhydrophilic (SH-PSpl150s) one (Fig. 4b). As a matter of fact, it is generally believed that hydrophilic substrates induce less modification in the conformation of adsorbed proteins; thus, proteins would retain a more active conformation on hydrophilic substrates as compared to the one that they adopt upon adsorption onto hydrophobic substrates.48 In the case of FN, several studies have confirmed that while hydrophilic surfaces promote the extension of its dimer arms, in a conformation that favors the binding of antibodies against cell-adhesive epitopes, hydrophobic surfaces promote the disruption of its secondary structures, negatively affecting cell response.28,33,49,50 However, these studies have been performed just on surfaces with wettabilities in the hydrophobic–hydrophilic range.
Generally, protein adsorption studies on superhydrophobic and/or superhydrophilic surfaces are meant to determine the effectiveness of such surfaces in resisting protein adsorption,51–55 and do not investigate their conformation. We have previously shown that FN adopts an altered conformation on rough SH-PS compared to smooth PS, and this phenomenon consequently affected cellular response.17 This conformational change was argued to be a consequence of the extreme wetting property of the substrate rather than a direct effect of surface roughness.17 Indeed, even though micro- and nanotopographical cues are known to influence protein adsorption, in terms of amount,56–58 conformation,59–61 and distribution,58 in the case of a SH substrate the roughness of the surface provokes specific phenomena at the material interface due to the Cassie–Baxter state: the suspension of the liquid over the asperities of the surface provokes the appearance of shear forces that affect the conformation of the protein,62 possibly leading to its denaturation. At increasing wettability, besides the already discussed positive effect of the surface intrinsic hydrophilicity, the transition from the Cassie–Baxter to the Wenzel state might also promote higher FN activity. Indeed, the penetration of the liquid into the asperities of the surface annuls the above-mentioned denaturing forces and simultaneously exposes the entire rough surface to the solution containing the adsorbing proteins; in this situation, the nanotopographical features of the roughness might favor FN adsorption in a conformation that enhances the exposition of cell-binding epitopes; a surface presenting 200 nm spherical features was in fact observed to induce FN spreading and consequently enhance cell adhesion.59 In our system, this interplay of surface nanotopography and surface energy appears to prompt an optimal FN conformation on the hydrophilic surface (SH-PSpl35s, WCA = 38.6 ± 8.1°).
2.3 Myogenic differentiation
Cell response to this family of substrates with wettability ranging from the superhydrophobic to the superhydrophilic regime was assessed by studying the myogenic differentiation of C2C12 cells. Sarcomeric myosin immunodetection revealed a non-monotonic dependence of the degree of differentiation on surface wettability, with a maximal enhancement of differentiation on the hydrophilic substrate (SH-PSpl35s) (Fig. 5). Specifically, on SH-PS myogenic differentiation was diminished with respect to the collagen type I control, considered as the standard substrate for myogenic differentiation;50,63 then, it progressively increased with increasing wettability until the hydrophilic surface, the increase between the samples and with respect to the control being statistically significant (Fig. 5b). Cell density was observed to increase substantially with increasing wettability, and to stabilize to similar values on the hydrophilic and superhydrophilic substrates (Fig. 5b). When cells were cultured along a gradient of wettability spanning the same range of wetting properties as the discrete samples, the same non-monotonic differentiation pattern was found, but the differences between the positions along the gradient were enhanced and, overall, the differentiation levels were significantly reduced with respect to the collagen control (Fig. 6). This differential behavior between discrete samples and gradient is explained in terms of the migration phenomena that occur when cells are exposed to a continuously changing range of stimuli, which will be further discussed later in this section.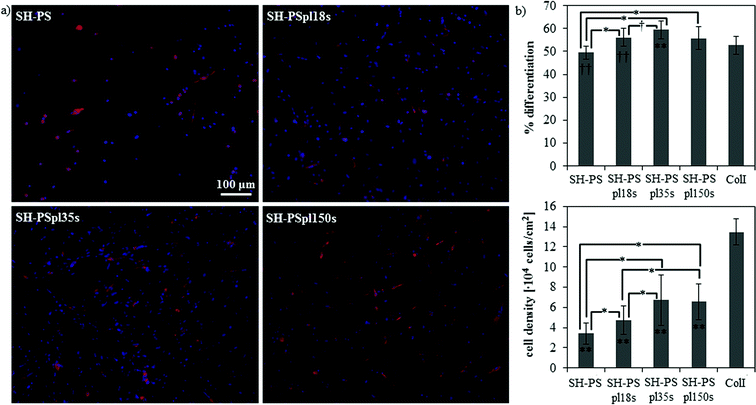 | ||
| Fig. 5 Myogenic differentiation on SH-PS and on Ar-plasma treated samples with different treatment times. (a) Fluorescence staining of sarcomeric myosin-positive cells and cell nuclei. (b) Myogenic differentiation (as determined by the percentage of sarcomeric myosin-positive cells) and cell density. ColI indicates the collagen type I control; statistically significant differences between the samples (as determined by ANOVA) are indicated with * (p < 0.05) and † (p < 0.1); statistically significant differences with the control are indicated with ** (p < 0.05) and †† (p < 0.1). | ||
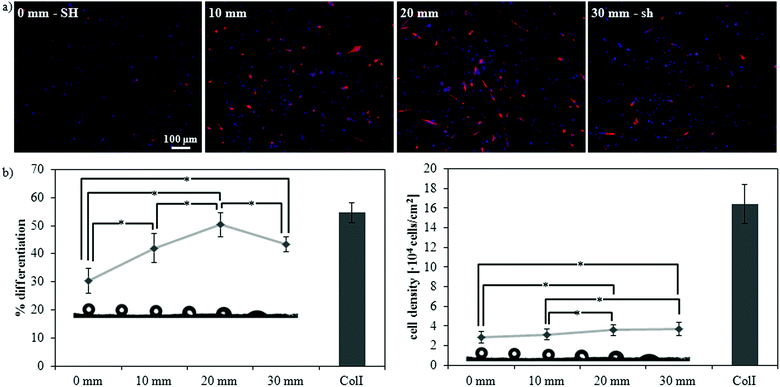 | ||
| Fig. 6 Myogenic differentiation along a gradient of wettability from superhydrophobicity (SH) to superhydrophilicity (sh). (a) Fluorescence staining of sarcomeric myosin-positive cells and cell nuclei at four different distances along the gradient: 0 mm (SH), 10 mm (hydrophobic, WCA ∼ 115°), 20 mm (hydrophilic, WCA ∼ 75°), and 30 mm (sh). (b) Myogenic differentiation (as determined by the percentage of sarcomeric myosin-positive cells) and cell density at different distances along the gradient. ColI indicates the collagen type I control; statistically significant differences between the samples (as determined by ANOVA) are indicated with * (p < 0.05); all samples are significantly different (p < 0.05) with respect to the control. | ||
On the whole, these differentiation results may be justified as a consequence of the different conformation that FN adopts on these surfaces (Fig. 4b). Both on the discrete samples and along the gradient a maximum of myogenic differentiation was encountered where FN activity was the highest, i.e., on the hydrophilic sample and in the hydrophilic portion of the gradient; in fact, a FN conformation that favors the exposure of α5β1 integrin binding sites has been shown to be essential to trigger myogenic differentiation.32,50,63 Within this perspective, our results suggest that the modulation of FN conformation may be used to tune myogenic differentiation. Moreover, it supports the hypothesis that initial events at the cell-material interface (e.g., protein adsorption and cell adhesion) determine cell functionality in the long term (e.g., cell differentiation).
Further insight on this phenomenon may be obtained by observing cell response along the gradient at different time points (3 h and 24 h) (Fig. 7). Initial cell adhesion was impaired on the SH end of the gradient, where fewer cells adhered and presented a not completely matured cytoskeleton with no evidence of defined stress fibers; at increasing hydrophilicities a higher cell density was detected and cells gradually adopted a more spread morphology and a more developed cytoskeleton organization with clear stress fibers (Fig. 7a–c). After 24 h of culture, cell density increased (Fig. 7a) and cells started to acquire a more mature morphology, with a well-developed actin cytoskeleton, also on the SH portion of the gradient (Fig. 7d and e). After the first day of culture, cell density increased only on the superhydrophobic and hydrophobic portions of the gradient, whilst it remained stable on the hydrophilic and superhydrophilic ones (Fig. 7a). On the whole, these data indicate a delayed cell response on the SH surface, confirming previous results;11,14,16,17 cells have been in fact shown to adhere mainly to some points of the asperities on the surface, since the Cassie–Baxter state prevents the medium and the cells from being in contact with the entire surface.11 As a result, cells adopt rounder morphologies11,17 and, if presented with patterned superhydrophobic/superhydrophilic substrates, adhere preferentially onto the wettable regions,14 where the medium completely wets the roughness of the surface. In time, cells adhere to the superhydrophobic substrate and start proliferating;14 a partial transition from the Cassie–Baxter to the Wenzel state might also occur during the culture,16 contributing to this late cell response. As a result, in our system the delay in cell adhesion on the superhydrophobic region correspondingly detains the myogenic differentiation process. On the other hand, on hydrophilic and superhydrophilic surfaces, cells have been shown to adhere promptly11,14 and, in these conditions, the differentiation process is not detained. Within the cell–protein–material interaction paradigm, the differential adhesion behavior observed on surfaces with distinct wettabilities is again a consequence of the ECM proteins adsorbed onto the substrate. In our case, the different conformation of the absorbed FN appears to influence initial cell adhesion in a way that strengthens the above mentioned effect that protein conformation has on myogenic differentiation.
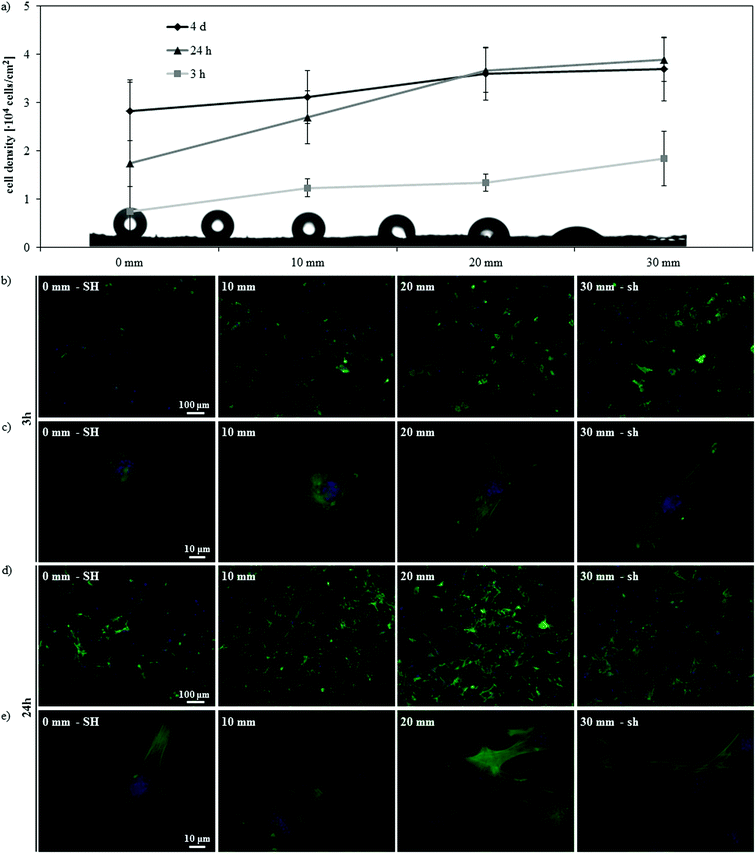 | ||
| Fig. 7 Effect of culturing time on the gradient surface. (a) Cell density at different distances along the SH-to-sh gradient after 3 h, 24 h and 4 days of culture. (b–e) Fluorescence microscopy of actin cytoskeleton and nuclei of C2C12 cells along the gradient after 3 h (b, c) and 24 h (d, e) of culture, at lower (b, d) and higher (c, e) magnification. | ||
Likewise, the community effect that may derive from higher cell densities might favor myogenic differentiation on areas with higher wettability, where the number of adhering cells is greater compared to substrates with lower wettability. Several authors have investigated the effect of cell density on cell differentiation.64–68 For the myogenic lineage, the ranges of densities considered in the literature span a ten-fold or hundred-fold variation between high and low cell density;64–67 that is a much greater range than the one encountered in our study (approximately a 2-fold and a 1.3-fold maximum variation between samples or portions within the gradients, respectively). On the other hand, Gobaa et al. reported a strong cell seeding density-dependence for the adipogenic differentiation of cells of the mesenchymal lineage at low density differences; however, the entire range of cell densities considered by the authors is lower than the one employed in our study.68 Hence, cell density, although possibly having a favorable effect on differentiation, is not deemed to be the driving force that determines cell response in the conditions of the present work. Within this respect, it is worth noticing that surfaces that promote similar cell attachment, such as the hydrophilic and superhydrophilic one (Fig. 5b and 6b), display a different behavior in terms of myogenic differentiation, as a result of the changes in protein conformation.
Along the gradient, where cells are exposed to a continuously changing range of stimuli, cell response differs from the one observed on the discrete uniform samples in two aspects: the diminished levels of differentiation and the enhancement of the differences in the degrees of differentiation between different wettabilities. These phenomena result from the interplay of migration and differentiation along the gradient, as previously observed by us for C2C12 cells cultured along a gradient of FN activity.4 Cell migration is in fact inversely correlated with the differentiation of myoblasts,69,70 so that migration phenomena along the gradient may delay and hamper cell differentiation with respect to discrete uniform samples. Cells, whichever portion along the gradient they initially adhere to, are in fact exposed to a continuous range of dynamically changing stimuli, so that the environment that they experience is intrinsically different than a uniform one and may locally trigger cell migration phenomena. The previously discussed differences in the velocity of initial cell adhesion on the various parts of the gradient might also play a role in determining these changes in cell differentiation levels. Cells would in fact initially concentrate on the portions of the gradient with higher FN activity, delaying further the response on the superhydrophobic surface and enhancing the differences in the differentiation levels between different wettabilities. Likewise, during the initial stages of culture, some of the cells seeded onto areas of lower FN activity may detach and adhere to portions of higher FN activity, contributing to the same effect of enhancement of the differences in the differentiation levels.
3. Experimental section
3.1 Preparation of the superhydrophobic surface
Superhydrophobic polystyrene (SH-PS) surfaces were obtained using a one-step phase-separation methodology under ambient atmosphere.35 A 70 mg mL−1 solution of polystyrene (PS, commercial granules of an injection molding grade, Sigma-Aldrich, St. Louis, Missouri, USA) was prepared in tetrahydrofuran (reagent A.C.S., Sigma-Aldrich); then, to each 2 mL of this solution 1.3 mL of ethanol (Scharlab, S.L., Sentmenat, Spain) were added and the solution was stirred. A few drops of the mixture were dipped onto a 0.25 mm-thick smooth PS film (Goodfellow Cambridge Ltd., Huntingdon, England); after 10 s, the substrate with the mixture was immersed in ethanol for 1 min. Finally, the surface was dried under a nitrogen flow and a rough superhydrophobic PS surface was obtained. The treated film was then cut into 9-mm-diameter circular or 5 × 30 mm2 rectangular samples for further processing, characterization, or use.3.2 Plasma treatment
To control surface wettability we employed a plasma discharge in an argon working atmosphere.11,35 The plasma treatment was carried out in a PICCOLO apparatus (Plasma Electronic GmbH, Neuenburg, Germany) for low pressure plasmas (10 to 100 Pa), which has a stainless steel vacuum chamber with a volume of 45 L. Plasma was generated using a 2.45 GHz-microwave system Gigatron®, with a discharge power up to 600 W. Concretely, the process started with the evacuation of the air present in the chamber until a base pressure of 18 Pa was achieved; after 15 s of homogenization with a controlled flow of argon (100 sccm), the plasma was generated at 30 W for a treatment time up to 150 s, inducing the oxidation of the samples and the consequent change of wettability.353.3 Preparation of the surface gradient
In order to obtain a gradient of wettability, a moving mask was used to allow a gradual exposition of 5 × 30 mm2 rectangular samples to the Ar-plasma (Fig. 3a). A stepper motor built in-house with commercial parts (LEGO Technic, The LEGO Group, Billund, Denmark) was introduced into the vacuum chamber and pulled the mask at a constant velocity of 140 μm s−1 during a treatment time of 70 s. The introduction of the motor inside the vacuum chamber forced the base pressure to increase to 99 Pa, in which conditions a plasma discharge of 70 s is sufficient to yield a superhydrophilic surface.3.4 Water contact angle
Static water contact angles (WCAs) were measured using the video-based optical contact angle measuring instrument OCA 20 (DataPhysics Instruments GmbH, Filderstadt, Germany) and water, reagent A.C.S. (Sigma-Aldrich), using the sessile drop method; the volume of the drop was 4 μL. The stability of the measurements was checked up to 30 d after the surface treatments.3.5 Scanning electron microscopy
The samples were sputter coated with gold and their morphology was analyzed with a JEOL JSM-6300 scanning electron microscope (SEM) (JEOL Ltd, Tokyo, Japan) via detection of the secondary electrons.3.6 Atomic force microscopy
AFM analyses were performed using a Multimode AFM equipped with a NanoScope IIIa controller (Veeco Instruments Inc., Plainview, New York, USA) operating in tapping mode in air; the Nanoscope 5.30r2 software version was used for image processing and analysis. Si-Cantilevers from Veeco were used, with a force constant of 2.8 N m−1 and a resonance frequency of 75 kHz. The phase signal was set to zero at a frequency 5–10% lower than the resonance one. Drive amplitude was 200 mV and the amplitude set-point (Asp) was 1.4 V. The ratio between the amplitude set-point and the free amplitude (Asp/A0) was kept equal to 0.7. Height, phase and amplitude magnitude were recorded simultaneously for each image. Height signals were used to calculate the roughness of the surfaces using the Nanoscope 5.30r2 software.3.7 Protein adsorption
Fibronectin (FN) from human plasma (Invitrogen Corp., Carlsbad, California, USA) was adsorbed on the different substrates by immersing them in a FN solution of concentration 20 μg mL−1 in Dulbecco's phosphate buffer saline (DPBS) (Sigma-Aldrich) for 1 h. After adsorption, samples were rinsed in DPBS to eliminate the non-adsorbed protein.3.8 Western blot
To quantify the amount of adsorbed fibronectin, we measured the protein remaining in the supernatant, i.e., the amount of protein that remained in solution after adsorption, as explained elsewhere.71 Briefly, different aliquots of non-adsorbed protein were subjected to 5% sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE), using Laemmli buffer 2× and standard denaturing conditions. Proteins were transferred to a positively charged polyvinylidene difluoride nylon membrane (GE Healthcare, Little Chalfont, United Kingdom) using a semi-dry transfer cell system (Bio-Rad Laboratories Inc., Hercules, California, USA), and blocked by immersion in 5% skimmed milk in PBS for 1 h at room temperature. The blot was incubated with anti-human fibronectin polyclonal antibody (developed in rabbit, Sigma-Aldrich) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) in PBS and washed three times (10 min each) with PBS containing 0.1% Tween 20 (Sigma-Aldrich) and 2% skimmed milk. The blot was subsequently incubated in horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G (GE Healthcare) diluted 1
500) in PBS and washed three times (10 min each) with PBS containing 0.1% Tween 20 (Sigma-Aldrich) and 2% skimmed milk. The blot was subsequently incubated in horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G (GE Healthcare) diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in PBS containing 0.1% Tween 20 and 2% skimmed milk (1 h at room temperature). The enhanced chemiluminescence detection system (GE Healthcare) was used according to the manufacturer's instructions prior to exposing the blot to an X-ray film for 1 min. Image analysis of the western bands was done using in-house software developed under MATLAB R2009b (The MathWorks Inc., Natick, Massachusetts, USA). All the western blotting bands were digitized using the same scanner (Epson Stylus Photo RX500, Seiko Epson Corp., Nagano, Japan) and the same scan parameters: 8 bits gray scale image and 300 dpi. The digitized images were binarized using the Otsu method, which chooses the threshold that minimizes the intraclass variance of the thresholded black and white pixels, in order to create a mask that automatically selected the edge of each western blot band.72 This mask was applied to a negative version of the original scanned picture providing a resulting image which contained only the western bands. The last step of the process consisted of adding all the pixels that conformed each band correctly weighted by their intensity level.
000 in PBS containing 0.1% Tween 20 and 2% skimmed milk (1 h at room temperature). The enhanced chemiluminescence detection system (GE Healthcare) was used according to the manufacturer's instructions prior to exposing the blot to an X-ray film for 1 min. Image analysis of the western bands was done using in-house software developed under MATLAB R2009b (The MathWorks Inc., Natick, Massachusetts, USA). All the western blotting bands were digitized using the same scanner (Epson Stylus Photo RX500, Seiko Epson Corp., Nagano, Japan) and the same scan parameters: 8 bits gray scale image and 300 dpi. The digitized images were binarized using the Otsu method, which chooses the threshold that minimizes the intraclass variance of the thresholded black and white pixels, in order to create a mask that automatically selected the edge of each western blot band.72 This mask was applied to a negative version of the original scanned picture providing a resulting image which contained only the western bands. The last step of the process consisted of adding all the pixels that conformed each band correctly weighted by their intensity level.
3.9 Enzyme-linked immunosorbent assay
After FN adsorption and rinsing in DPBS, surfaces were blocked against non-specific antibody binding using a blocking buffer (1% BSA–DPBS) for 30 min at 37 °C. They were then were incubated in the primary monoclonal antibody HFN7.1 (Developmental Studies Hybridoma Bank, Iowa City, Iowa, USA), which is directed against the flexible linker between the 9th and 10th type III repeat,47 with a dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000 for 1 h at 37 °C. After washing with 0.1% Tween 20–DPBS, substrates were incubated in alkaline phosphatase conjugated anti-mouse IgG (1
4000 for 1 h at 37 °C. After washing with 0.1% Tween 20–DPBS, substrates were incubated in alkaline phosphatase conjugated anti-mouse IgG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) for 1 h at 37 °C (Jackson ImmunoResearch Laboratories Inc., West Grove, Pennsylvania, USA), washed again, and incubated in 4-methylumbelliferyl phosphate (4-MUP) liquid substrate system (Sigma-Aldrich) for 45 min at 37 °C. Reaction products were quantified using a fluorescence plate reader (Victor3, PerkinElmer, Waltham, Massachusetts) at 365 nm excitation/460 nm emission. The background signal for each sample type was obtained by performing the same assay without previous protein adsorption onto the sample.
5000) for 1 h at 37 °C (Jackson ImmunoResearch Laboratories Inc., West Grove, Pennsylvania, USA), washed again, and incubated in 4-methylumbelliferyl phosphate (4-MUP) liquid substrate system (Sigma-Aldrich) for 45 min at 37 °C. Reaction products were quantified using a fluorescence plate reader (Victor3, PerkinElmer, Waltham, Massachusetts) at 365 nm excitation/460 nm emission. The background signal for each sample type was obtained by performing the same assay without previous protein adsorption onto the sample.
3.10 Cell cultures
Murine C2C12 myoblasts were obtained from ATCC (American Type Culture Collection, Manassas, Virginia, USA). Cells were maintained in Dulbecco's modified Eagle medium (DMEM) (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) supplemented with 20% fetal bovine serum (Thermo Fisher Scientific Inc.) and 1% penicillin–streptomycin (Lonza, Basel, Switzerland) and passaged twice a week using standard techniques. The FN-coated samples were seeded with 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells cm−2 in DMEM supplemented with 1% penicillin–streptomycin and 1% insulin–transferrin–selenium-X (Invitrogen Corp.) to induce myogenic differentiation, and maintained at 37 °C in a humidified atmosphere with 5% CO2 for 4 days; the culture medium was changed at the end of the second day of culture. As controls, cover glasses coated with a 1 mg mL−1 collagen I solution (Collagen Solution, STEMCELL Technologies Inc., Vancouver, Canada) and subsequently adsorbed with FN from a 10 μg mL−1 solution were used.
000 cells cm−2 in DMEM supplemented with 1% penicillin–streptomycin and 1% insulin–transferrin–selenium-X (Invitrogen Corp.) to induce myogenic differentiation, and maintained at 37 °C in a humidified atmosphere with 5% CO2 for 4 days; the culture medium was changed at the end of the second day of culture. As controls, cover glasses coated with a 1 mg mL−1 collagen I solution (Collagen Solution, STEMCELL Technologies Inc., Vancouver, Canada) and subsequently adsorbed with FN from a 10 μg mL−1 solution were used.
3.11 Immunohistochemistry assays
C2C12 cells were cultured on FN-coated materials for 4 days under differentiation conditions and then immunostained for sarcomeric myosin. Briefly, cultures were washed in DPBS, fixed in 70% ethanol–37% formaldehyde–glacial acetic acid (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and then blocked in 5% goat serum in DPBS for 1 h. Samples were sequentially incubated in MF-20 mouse antibody (Developmental Studies Hybridoma Bank) 1
1) and then blocked in 5% goat serum in DPBS for 1 h. Samples were sequentially incubated in MF-20 mouse antibody (Developmental Studies Hybridoma Bank) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 and anti-mouse Cy3-conjugated secondary antibody (Jackson Immunoresearch) 1
250 and anti-mouse Cy3-conjugated secondary antibody (Jackson Immunoresearch) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200. Finally, samples were washed before being mounted in Vectashield containing DAPI (Vector Laboratories, Inc., Burlingame, California, USA).
200. Finally, samples were washed before being mounted in Vectashield containing DAPI (Vector Laboratories, Inc., Burlingame, California, USA).
For the analysis of cell density and cell morphology along the wettability gradient at different time points (3 h, 24 h), cells were washed in DPBS and fixed in a 10% formalin solution (Sigma-Aldrich) at 4 °C for 30 min. Samples were then rinsed with DPBS and maintained in a permeabilizing buffer (103 g L−1 sucrose, 2.92 g L−1 NaCl, 0.6 g L−1 MgCl2, 4.76 g L−1 HEPES buffer, 5 mL L−1 Triton X-100, pH 7.2) at room temperature for 5 min. Afterwards, samples were incubated with BODIPY FL phallacidin (Invitrogen Corp.) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 at room temperature for 1 h. Finally, samples were washed twice in 0.5% Tween 20–DPBS before mounting with Vectashield containing DAPI (Vector Laboratories Inc.).
40 at room temperature for 1 h. Finally, samples were washed twice in 0.5% Tween 20–DPBS before mounting with Vectashield containing DAPI (Vector Laboratories Inc.).
A Nikon Eclipse 80i fluorescent microscope (Nikon Corporation, Tokyo, Japan) was used for cellular imaging. Cell density and differentiation were calculated by counting cell nuclei and the percentage of cells positive for sarcomeric myosin, respectively. Results were obtained by averaging the observations in three randomly selected fields for each one of the three replicas of every sample type. The CellC software was used for this purpose, after validation via comparison of software counts with manual enumeration of at least three images for each sample type.73
3.12 Statistics
Unless otherwise specified, each experiment was performed at least in triplicate, and the values of the measurements were reported as the average of the samples, with the variability expressed in terms of standard deviation. Statistically significant differences were determined by one-way ANOVA, with p < 0.05 or 0.1.4. Conclusions
In this work, we have produced a family of surfaces with hierarchical roughness at the micro and nanoscale that, thanks to a plasma surface treatment at increasing times, span the entire range of wettability, from the superhydrophobic to the superhydrophilic regime. Likewise, we have employed the same methodology to achieve a superhydrophobic-to-superhydrophilic surface gradient. To our knowledge, this is the first study where protein adsorption, not only in terms of amount, but also of conformation, and cell response, particularly in terms of a higher order cellular activity, such as differentiation, have been characterized along this entire range of wettabilities.Specifically, these surfaces modulate fibronectin conformation: the availability of cell-adhesive epitopes exhibits a non-monotonic dependence on wettability, with a higher fibronectin activity on the hydrophilic substrate. On the other hand, the exposure of cell-binding sites is diminished on the surfaces with extreme wetting properties, the conformation being particularly altered on the superhydrophobic substrate. C2C12 cells respond accordingly to the fibronectin-coated substrates: the hydrophilic surface supports enhanced myogenic differentiation, both on the discrete uniform samples and along the gradient; moreover, on the superhydrophobic surface the altered fibronectin conformation delays cell adhesion, further retaining the myogenic differentiation process. Finally, the analysis of the cell response along the superhydrophobic-to-superhydrophilic gradient reveals that the exposure of the cells to a continuous range of dynamically changing stimuli alters cell behavior in a way that hampers the overall levels of differentiation and enhances the differential response, as measured by the degree of myogenic differentiation, to the different wettabilities.
Acknowledgements
The support of the Spanish Ministry of Science and Innovation through project MAT2009-14440-C02-01 is acknowledged. CIBER-BBN is an initiative funded by the VI National R&D&I Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund.Notes and references
- M. Singh, C. Berkland and M. S. Detamore, Tissue Eng., Part B: Rev., 2008, 14, 341–366 CrossRef CAS.
- S. B. Kennedy, N. R. Washburn, C. G. Simon and E. J. Amis, Biomaterials, 2006, 27, 3817–3824 CrossRef CAS.
- J. R. Tse and A. J. Engler, PLoS One, 2011, 6, e15978 CAS.
- J. Ballester-Beltrán, M. Cantini, M. Lebourg, P. Rico, D. Moratal, A. J. García and M. Salmerón-Sánchez, J. Mater. Sci.: Mater. Med., 2012, 23, 195–204 CrossRef.
- M. S. Kim, G. Khang and H. B. Lee, Prog. Polym. Sci., 2008, 33, 138–164 CrossRef CAS.
- J. Zhang and Y. Han, Langmuir, 2008, 24, 796–801 CrossRef CAS.
- M. K. Chaudhury and G. M. Whitesides, Science, 1992, 256, 1539–1541 CAS.
- T. G. Ruardy, J. M. Schakenraad, H. C. van der Mei and H. J. Busscher, Surf. Sci. Rep., 1997, 29, 3–30 CrossRef.
- M. Zelzer, R. Majani, J. W. Bradley, F. R. A. J. Rose, M. C. Davies and M. R. Alexander, Biomaterials, 2008, 29, 172–184 CrossRef CAS.
- J. Yang, F. R. A. J. Rose, N. Gadegaard and M. R. Alexander, Adv. Mater., 2009, 21, 300–304 CrossRef CAS.
- W. Song, D. D. Veiga, C. A. Custódio and J. F. Mano, Adv. Mater., 2009, 21, 1830–1834 CrossRef CAS.
- X. Yu, Z. Wang, Y. Jiang and X. Zhang, Langmuir, 2006, 22, 4483–4486 CrossRef CAS.
- T. Sun, H. Tan, D. Han, Q. Fu and L. Jiang, Small, 2005, 1, 959–963 CrossRef CAS.
- T. Ishizaki, N. Saito and O. Takai, Langmuir, 2010, 26, 8147–8154 CrossRef CAS.
- A. I. Neto, C. A. Custódio, W. Song and J. F. Mano, Soft Matter, 2011, 7, 4147 RSC.
- S. M. Oliveira, W. Song, N. M. Alves and J. F. Mano, Soft Matter, 2011, 7, 8932–8941 RSC.
- J. Ballester-Beltrán, P. Rico, D. Moratal, W. Song, J. F. Mano and M. Salmerón-Sánchez, Soft Matter, 2011, 7, 10803–10811 RSC.
- X. Li, H. Dai, S. Tan, X. Zhang, H. Liu, Y. Wang, N. Zhao and J. Xu, J. Colloid Interface Sci., 2009, 340, 93–97 CrossRef CAS.
- Y.-H. Lai, J.-T. Yang and D.-B. Shieh, Lab Chip, 2010, 10, 499–504 RSC.
- A. J. García, Adv. Polym. Sci., 2006, 203, 171–190 CrossRef.
- B. M. Gumbiner, Cell, 1996, 84, 345–357 CrossRef CAS.
- C. Werner, T. Pompe and P. Katrin, Adv. Polym. Sci., 2006, 203, 63–93 CrossRef CAS.
- M. Salmerón-Sánchez and G. Altankov, in Tissue Engineering, ed. D. Eberli, In-Tech, 2010, vol. 1, pp. 77–102 Search PubMed.
- R. O. Hynes, Cell, 2002, 110, 673–687 CrossRef CAS.
- B. Geiger, A. Bershadsky, R. Pankov and K. M. Yamada, Nat. Rev. Mol. Cell Biol., 2001, 2, 793–805 CrossRef CAS.
- E. Pearlstein, L. I. Gold and A. Garcia-Pardo, Mol. Cell. Biochem., 1980, 29, 103–128 CrossRef CAS.
- B. G. Keselowsky, D. M. Collard and A. J. García, J. Biomed. Mater. Res., 2003, 66A, 247–259 CrossRef CAS.
- K. E. Michael, V. N. Vernekar, B. G. Keselowsky, J. C. Meredith, R. A. Latour and A. J. García, Langmuir, 2003, 19, 8033–8040 CrossRef CAS.
- A. J. García and D. Boettiger, Biomaterials, 1999, 20, 2427–2433 CrossRef.
- G. K. Toworfe, R. J. Composto, C. S. Adams, I. M. Shapiro and P. Ducheyne, J. Biomed. Mater. Res., 2004, 71A, 449–461 CrossRef CAS.
- L. Baugh and V. Vogel, J. Biomed. Mater. Res., 2004, 69A, 525–534 CrossRef CAS.
- M. A. Lan, C. A. Gersbach, K. E. Michael, B. G. Keselowsky and A. J. García, Biomaterials, 2005, 26, 4523–4531 CrossRef CAS.
- F. Grinnell and M. K. Feld, J. Biol. Chem., 1982, 257, 4888–4893 CAS.
- G. Altankov, V. Thom, T. Groth, K. Jankova, G. Jonsson and M. Ulbricht, J. Biomed. Mater. Res., 2000, 52, 219–230 CrossRef CAS.
- N. M. Oliveira, A. I. Neto, W. Song and J. F. Mano, Appl. Phys. Express, 2010, 3, 085205 CrossRef.
- A. B. D. Cassie and S. Baxter, Trans. Faraday Soc., 1944, 40, 546–551 RSC.
- X.-M. Li, D. Reinhoudt and M. Crego-Calama, Chem. Soc. Rev., 2007, 36, 1350–1368 RSC.
- J. Bico, C. Tordeux and D. Quéré, Europhys. Lett., 2001, 55, 214–220 CrossRef CAS.
- G. McHale, N. J. Shirtcliffe, S. Aqil, C. C. Perry and M. I. Newton, Phys. Rev. Lett., 2004, 93, 036102 CrossRef CAS.
- R. N. Wenzel, Ind. Eng. Chem., 1936, 28, 988–994 CrossRef CAS.
- J. D. Sipe, Ann. N. Y. Acad. Sci., 2002, 961, 1–9 CrossRef CAS.
- L. G. Griffith and G. Naughton, Science, 2002, 295, 1009–1014 CrossRef CAS.
- F. Grinnell, J. Cell Biol., 1986, 103, 2697–2706 CrossRef CAS.
- D. J. Iuliano, S. S. Saavedra and G. A. Truskey, J. Biomed. Mater. Res., 1993, 27, 1103–1113 CrossRef CAS.
- T. P. Ugarova, C. Zamarron, Y. Veklich, R. D. Bowditch, M. H. Ginsberg, J. W. Weisel and E. F. Plow, Biochemistry, 1995, 34, 4457–4466 CrossRef CAS.
- K. B. Mcclary, T. Ugarova and D. W. Grainger, J. Biomed. Mater. Res., 2000, 50, 428–439 CrossRef CAS.
- R. C. Schoen, K. L. Bentley and R. J. Klebe, Hybridoma, 1982, 1, 99–108 CrossRef CAS.
- K. Anselme, A. Ponche and M. Bigerelle, Proc. Inst. Mech. Eng., Part H, 2010, 224, 1487–1507 CrossRef CAS.
- S. Cheng, K. K. Chittur, C. N. Sukenik, L. A. Culp and K. Lewandowska, J. Colloid Interface Sci., 1994, 162, 135–143 CrossRef CAS.
- A. J. García, M. D. Vega and D. Boettiger, Mol. Biol. Cell, 1999, 10, 785–798 Search PubMed.
- R. B. Pernites, C. M. Santos, M. Maldonado, R. R. Ponnapati, D. F. Rodrigues and R. C. Advincula, Chem. Mater., 2012, 24, 870–880 CrossRef CAS.
- J.-Y. Shiu and P. L. Chen, Adv. Funct. Mater., 2007, 17, 2680–2686 CrossRef CAS.
- K. Tsougeni, P. S. Petrou, D. P. Papageorgiou, S. E. Kakabakos, A. Tserepi and E. Gogolides, Sens. Actuators, B, 2012, 161, 216–222 CrossRef CAS.
- S. Gam-derouich, M. Gosecka, S. Lepinay, M. Turmine, B. Carbonnier, T. Basinska, S. Slomkowski, M.-C. Millot, A. Othmane, D. B. Hassen-chehimi and M. M. Chehimi, Langmuir, 2011, 27, 9285–9294 CrossRef CAS.
- P. Patel, C. K. Choi and D. D. Meng, J. Assoc. Lab. Autom., 2009, 15, 114–119 CrossRef.
- M. N. Sela, L. Badihi, G. Rosen, D. Steinberg and D. Kohavi, Clin. Oral Implants Res., 2007, 18, 630–638 CrossRef.
- D. Khang, S. Y. Kim, P. Liu-Snyder, G. T. R. Palmore, S. M. Durbin and T. J. Webster, Biomaterials, 2007, 28, 4756–4768 CrossRef CAS.
- C. González-García, S. R. Sousa, D. Moratal, P. Rico and M. Salmerón-Sánchez, Colloids Surf., B, 2010, 77, 181–190 CrossRef.
- D. C. Miller, K. M. Haberstroh and T. J. Webster, J. Biomed. Mater. Res., Part A, 2007, 81A, 678–684 CrossRef CAS.
- E. Costa Martínez, J. C. Rodríguez Hernández, M. Machado, J. F. Mano, J. L. Gómez Ribelles, M. Monleón Pradas and M. Salmerón Sánchez, Tissue Eng. A, 2008, 14, 1751–1762 CrossRef.
- M. S. Lord, B. G. Cousins, P. J. Doherty, J. M. Whitelock, A. Simmons, R. L. Williams and B. K. Milthorpe, Biomaterials, 2006, 27, 4856–4862 CrossRef CAS.
- J. Ulmer, B. Geiger and J. P. Spatz, Soft Matter, 2008, 4, 1998–2007 RSC.
- M. Salmerón-Sánchez, P. Rico, D. Moratal, T. T. Lee, J. E. Schwarzbauer and A. J. García, Biomaterials, 2011, 32, 2099–2105 CrossRef.
- C. Lindon, O. Albagli, C. Pinset and D. Montarras, Dev. Biol., 2001, 240, 574–584 CrossRef CAS.
- K. Tanaka, K. Sato, T. Yoshida, T. Fukuda, K. Hanamura, N. Kojima, T. Shirao, T. Yanagawa and H. Watanabe, Muscle & Nerve, 2011, 44, 968–977 CrossRef CAS.
- S. R. Chowdhury, Y. Muneyuki, Y. Takezawa, M. Kino-oka, A. Saito, Y. Sawa and M. Taya, J. Biosci. Bioeng., 2010, 109, 310–313 CrossRef CAS.
- P. Kaspar, P. Pajer, D. Sedlak, T. Tamaoki and M. Dvorak, Exp. Cell Res., 2005, 309, 419–428 CrossRef CAS.
- S. Gobaa, S. Hoehnel, M. Roccio, A. Negro, S. Kobel and M. P. Lutolf, Nat. Methods, 2011, 8, 949–955 CrossRef CAS.
- B. A. Bondesen, K. A. Jones, W. C. Glasgow and G. K. Pavlath, FASEB J., 2007, 21, 3338–3345 CrossRef CAS.
- H. C. Olguin, C. Santander and E. Brandan, Dev. Biol., 2003, 259, 209–224 CrossRef CAS.
- P. Rico, J. C. Rodríguez Hernández, D. Moratal, G. Altankov, M. Monleón Pradas and M. Salmerón-Sánchez, Tissue Eng. A, 2009, 15, 3271–3281 CrossRef CAS.
- N. Otsu, IEEE Trans. Syst. Man Cybern., 1979, 9, 62–66 CrossRef.
- J. Selinummi, J. Seppälä, O. Yli-Harja and J. Puhakka, BioTechniques, 2005, 39, 859–863 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work |
| This journal is © The Royal Society of Chemistry 2013 |
