The toxic effect of silver ions and silver nanoparticles towards bacteria and human cells occurs in the same concentration range
Christina
Greulich
a,
Dieter
Braun
b,
Alexander
Peetsch
c,
Jörg
Diendorf
c,
Bettina
Siebers
*b,
Matthias
Epple
*c and
Manfred
Köller
*a
aBergmannsheil University Hospital/Surgical Research, Ruhr-University of Bochum, Buerkle-de-la-Camp-Platz 1, 44789, Bochum, Germany. E-mail: manfred.koeller@rub.de
bMolecular Enzyme Technology and Biochemistry, Biofilm Centre, University of Duisburg-Essen, Universitaetsstr. 5, 45141, Essen, Germany. E-mail: bettina.siebers@uni-due.de
cInorganic Chemistry and Center for Nanointegration Duisburg-Essen (CeNIDE), University of Duisburg-Essen, Universitaetsstr. 5-7, 45117, Essen, Germany. E-mail: matthias.epple@uni-due.de
First published on 13th June 2012
Abstract
Silver is commonly used both in ionic form and in nanoparticulate form as a bactericidal agent. This is generally ascribed to a higher toxicity towards prokaryotic cells than towards mammalian cells. Comparative studies with both silver ions (such as silver acetate) and polyvinylpyrrolidone (PVP)-stabilized silver nanoparticles (70 nm) showed that the toxic effect of silver occurs in a similar concentration range for Escherichia coli, Staphylococcus aureus, human mesenchymal stem cells (hMSCs), and peripheral blood mononuclear cells (PBMCs), i.e. 0.5 to 5 ppm for silver ions and 12.5 to 50 ppm for silver nanoparticles. For a better comparison, bacteria were cultivated both in Lysogeny broth medium (LB) and in Roswell Park Memorial Institute medium (RPMI)/10% fetal calf serum (FCS) medium, as the state of silver ions and silver nanoparticles may be different due to the presence of salts, and biomolecules like proteins. The effective toxic concentration of silver towards bacteria and human cells is almost the same.
Introduction
Silver is used in many consumer commodities and biomedical applications due to its antibacterial effect.1–3 It is a generally accepted assumption that silver is more toxic towards prokaryotes than towards mammalian cells, leading to a therapeutic window where mammalian tissue is not harmed but where bacteria are killed.2–9 In the biomedical field, catheter and implant coatings, bone cement, and sutures with silver compounds, e.g. silver salts, were proposed for antibacterial applications.10–12In addition to silver salts, silver nanoparticles are increasingly used due to their slower dissolution rate, leading to a continuous release of silver ions.9,13–17 The comparison of the biological effects of silver ions and silver nanoparticles is of high current interest because of the increasing application of silver nanoparticles in consumer products and medical products and the subsequent pathways in nature, e.g. in and after sewage plants.3,18,19
Silver ions have been reported to interact with a variety of biomolecules within a cell such as nucleic acids,20 cell wall components, sulfhydryl groups of metabolic enzymes,12,21,22 and sulfur-containing cell components like glutathione.23 In general, the toxicity of silver nanoparticles appears to be driven by the release of silver ions. The generation of reactive oxygen species (ROS) upon exposure of cells to nanoparticles is a major contributor to the toxicity of silver nanoparticles.24
Studies where the same silver species were analyzed under comparable growth conditions with respect to the cell culture medium in bacteria and mammalian cells are rare25 and sometimes carried out with unknown silver concentrations in the presence of a silver-releasing surface.26–28
We have therefore prepared well-defined silver-containing samples, i.e. silver acetate as a source of silver ions and PVP-stabilized silver nanoparticles which were well dispersed in cell culture media.29 The biological effect was tested on two bacterial strains, i.e. Escherichia coli DH5α (DSMZ 6897, gram-negative) and Staphylococcus aureus (DSMZ 1104, gram-positive) as well as three mammalian cell types, i.e. human mesenchymal stem cells (hMSC) and human peripheral blood mononuclear cells (PBMC), which mainly consist of lymphocytes (e.g. T-cells) and monocytes. To achieve a better comparison with in vivo conditions, the bacteria were cultured both in bacterial growth medium and in cell culture medium.
Therefore, the results permit a direct comparison of the toxic action of silver in the form of ions and in the form of nanoparticles towards bacteria and human cells.
Experimental
Preparation of silver acetate solutions
Silver acetate (Sigma-Aldrich; ReagentPlus®, 99%) was dissolved in ultrapure water, prepared with an ELGA PURELAB Ultra (ELGA Labwater, Germany) instrument.Preparation of silver nanoparticles
Silver nanoparticles were prepared as reported earlier by reduction of silver nitrate with glucose in the presence of poly(N-vinyl pyrrolidone), PVP.30,31 The dispersion was purified by ultracentrifugation, followed by redispersion in ultrapure water under ultrasonication. Therefore, neither dissolved silver ions nor excess polymer or glucose oxidation products were present. The final silver content was determined by atomic absorption spectroscopy (AAS; Thermo Electron Corporation, MSeries). The hydrodynamic diameter and the zeta potential of the dispersed particles were measured by dynamic light scattering (DLS) with a Malvern Zetasizer Nano ZS (Malvern Instruments, UK). Scanning electron microscopy (SEM) was carried out with a FEI Quanta 400 ESEM instrument (FEI, USA).PVP (K 30, Povidone 30; Fluka, molecular weight 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), silver nitrate (Fluka, p.a.), and D-(+)-glucose (Baker) were used for the synthesis of the silver nanoparticles. Ultrapure water was prepared with an ELGA PURELAB Ultra instrument.
000 g mol−1), silver nitrate (Fluka, p.a.), and D-(+)-glucose (Baker) were used for the synthesis of the silver nanoparticles. Ultrapure water was prepared with an ELGA PURELAB Ultra instrument.
Preparation of stock and working solutions
PVP-Coated spherical silver nanoparticles were dispersed in sterile ultrapure water at 1 g L−1 (1,000 ppm) as stock solution. A solution of silver acetate with 1 g L−1 was the second stock solution. Both stock solutions were diluted with ultrapure water to prepare working stock solutions for the individual experiments at different silver concentrations (10–1,000 ppm). The final concentrations were achieved by the addition of 50 μL of either silver nanoparticle or of silver acetate working stock solutions to a 1 mL cell suspension prior to cultivation. Therefore the concentrations of other medium ingredients were kept constant in all experiments.The final concentrations used for the bacterial tests were 50 ppm, 40 ppm, 30 ppm, 25 ppm, 20 ppm, and 12.5 ppm for silver nanoparticles, and 10 ppm, 7.5 ppm, 5 ppm, 3.5 ppm, 2.5 ppm, 1.25 ppm, and 1.0 ppm for silver ions. For the cell cultures, we used 50 ppm, 30 ppm, 20 ppm, 10 ppm, and 5 ppm for silver nanoparticles and 5 ppm, 2.5 ppm, 1.5 ppm, 1.0 ppm, and 0.5 ppm for silver ions. The nanoparticles were freshly prepared before the biological tests and stored under argon to avoid oxidation and the subsequent release of silver ions.13–16,32
All the concentration data given in the Tables and Figures refer to the mass of silver in the solution, regardless of its state (ions or nanoparticles).
Bacterial culture
Cell tests were performed with Escherichia coli DH5α (DSMZ 6897) and Staphylococcus aureus (DSMZ 1104) obtained from the DSMZ (German Collection of Microorganisms and Cell Cultures) and Life Technologies GmbH (Karlsruhe, Germany). Bacterial concentrations of overnight cultures were measured using a Densichek® (bioMérieux, Lyon, France) turbidity photometer. The calculation of bacterial counts was based on turbidity standard solutions (McFarland scale).The antimicrobial activity of ionic and nanoparticulate silver was tested using standard methods which determine the minimum inhibitory concentration (MIC) and the minimum bactericidal concentration (MBC). MIC was determined in two different liquid culture media: Lysogeny broth (LB) and RPMI 1640 (Life Technologies) containing 10% (v/v) fetal calf serum (FCS, Life Technologies) and L-glutamine (0.3 g L−1, Life Technologies), and defined as the lowest silver concentration able to inhibit bacterial growth (no visible growth) in 2 mL plastic macrodilution test tubes. Therefore, working silver stock solutions (50 μL) were added to 1 mL of the respective liquid culture medium, and different cell numbers (103 to 106 mL−1) of bacteria were used for inoculation. Cells were either incubated in a cell culture incubator (RPMI/10% FCS) in the presence of 5% carbon dioxide in a humidified atmosphere or in a microbiological incubator (LB broth) each at 37 °C overnight. To confirm the growth of the bacterial strains in both media, the cells were grown either in RPMI/10% FCS in a cell culture incubator or in LB broth in a microbial incubator in glass culture tubes containing 3 mL of the respective medium. The optical density was determined at 600 nm in a spectrophotometer (BioPhotometer plus, Eppendorf). Due to the turbidity induced by the silver nanoparticles, the MIC determination was possible only for the silver acetate samples.
The minimum bactericidal concentration (MBC) was subsequently determined by plating 100 μL aliquots of the MIC samples on agar plates (blood agar for S.aureus and LB agar for E. coli). The MBC was defined as the lowest silver concentration which completely prevented colony forming units (CFU) on the agar plate.
Cell culture
Human mesenchymal stem cells (hMSCs, 3rd to 7th passage, Lonza, Walkersville Inc., MD, USA) were cultured in cell culture medium RPMI/10% FCS using 24-well cell culture plates (Falcon, Becton Dickinson GmbH, Heidelberg, Germany). Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. hMSCs were sub-cultivated every 7–14 days depending on the cell proliferation. Adherent cells were washed with phosphate-buffered saline solution (PBS, GIBCO, Life Technologies) and detached from the culture flasks by addition of 0.2 mL cm−2 0.25% trypsin–0.1% ethylenediamine tetraacetic acid (EDTA, Sigma-Aldrich, Taufkirchen, Germany) for 5 min at 37 °C. Subsequently, the hMSCs were collected and washed twice with RPMI/10% FCS. Subconfluently growing hMSCs were incubated at 37 °C in the presence or absence of different concentrations of silver nanoparticles or silver ions for 24 h under cell culture conditions.The viability and the morphology of the incubated hMSCs were analyzed using calcein-acetoxymethylester (calcein-AM, Calbiochem, Schwalbach, Germany) fluorescence staining. After incubation for 24 h, the nanoparticle-treated and silver ion-treated cells were washed twice with RPMI and incubated with calcein-AM (1 μM) at 37 °C for 30 min under cell culture conditions. Subsequently, the adherent cells were washed again with RPMI and analyzed by fluorescence microscopy (Olympus MVX10, Olympus, Hamburg, Germany). Fluorescence microphotographs were taken (Cell P, Olympus) and digitally processed using Adobe Photoshop® 7.0. The cell morphology was analyzed by phase-contrast microscopy (Olympus CK 2) and documented by microphotography (Camedia C3030, Olympus). In this method, living cells give rise to green fluorescence.
PBMCs were isolated by a single-step procedure that was based on a discontinuous double-Ficoll gradient described by English and Andersen.33 Briefly, EDTA-anticoagulated peripheral blood (9 mL Monovette®, Sarstedt, Nürnbrecht, Germany) obtained from healthy volunteers (covered by the approval of the local ethics committee #3036-07) were diluted with an equal volume of 0.9% NaCl and carefully overlaid on a double gradient formed by layering 10 mL of aqueous polysucrose–sodium diatrizoate, adjusted to a density of 1.077 g mL−1 (Histopaque 1077, Sigma-Aldrich, Taufkirchen, Germany), on 10 mL Histopaque 1119 (Sigma-Aldrich) in 50 mL falcon tubes (BD Biosciences, Heidelberg, Germany). The tubes were centrifuged at 700 g for 30 min at room temperature. After centrifugation, two distinct leukocyte cell layers (PBMC and polymorphonuclear neutrophil granulo (PMN)) were obtained above the bottom sediment of erythrocytes. The PBMC layer was carefully aspirated and transferred to a fresh 50 mL falcon tube filled with phosphate buffered saline (PBS, Sigma-Aldrich) and centrifuged at 200 g for 15 min at 4 °C. This method led to more than 95% pure and viable PBMCs. Cell counting was performed using Tuerk staining solution (Sigma-Aldrich). The isolated cells were adjusted to 1 × 106 cells mL−1 in RPMI cell culture medium supplemented with L-glutamine (0.3 g L−1), sodium bicarbonate (2.0 g L−1), 10% (v/v) FCS, and 20 mM N-(2-hydroxy)-piperazine-N'-(2-ethanesulfonic acid) (HEPES, Sigma-Aldrich).
Freshly isolated PBMCs were distributed in each well (1 × 106 cells mL−1) of a 24-well culture plate and cultured at 37 °C for 24 h in the presence or absence of different concentrations of freshly prepared silver nanoparticles or silver acetate in a humidified atmosphere of 5% CO2. The viability of the incubated PMBCs was determined by analyzing the cell numbers by counting cell events for 30 s under constant flow using flow cytometry (FACSCalibur™, Becton Dickinson). Non-viable cells were excluded by 7-AAD (7-amino-actinomycin D, BD Biosciences, Heidelberg, Germany) fluorescence staining.
Statistical analysis
Data are expressed as mean ± standard deviation of at least three independent experiments. Analysis of the data distribution was performed using the Student's t-test to analyze the significance of differences between the treated group and the control group (without silver exposure). P values of less than 0.05 were considered to be statistically significant.Results and discussion
Silver acetate (ionic silver) and PVP-functionalized silver nanoparticles were used. Scanning electron microscopy showed that the silver nanoparticles had a spherical shape with a diameter of the metallic core of 70 ± 20 nm. The hydrodynamic diameter of the nanoparticles was 75 ± 20 nm, as measured by dynamic light scattering. The zeta-potential was −25 mV. The polydispersity index (PDI) was below 0.3 in all cases, indicating the absence of larger agglomerates. The silver nanoparticles were stored under argon to prevent partial oxidative dissolution (which drastically influences the nanoparticle toxicity15) before the cell culture experiments. Two bacterial strains, i.e. gram-negative E. coli and gram-positive S. aureus, and three human cell types, i.e. mesenchymal stem cells, T-lymphocytes, and monocytes, were subjected to silver in ionic and nanoparticulate form. As far as possible, the incubation conditions were the same in order to make the effects of the medium (e.g., a complexation or reduction of silver ions or a coating of silver nanoparticles with proteins) the same in all the experiments.15Bacteria and mammalian cells are usually cultivated in different media. However, for a strict comparison, we have used RPMI 1640 with 10% (v/v) FCS as a culture medium in both cases. This medium is typically used for the cultivation of mammalian cells and represents a good approximation of the in vivo conditions where the antibacterial action is desired. We have shown earlier that PVP-stabilized silver nanoparticles remain well dispersed in this medium.29 The bacteria were also cultivated in standard bacterial growth medium (LB) for comparison. Note that in LB medium, the silver nanoparticles were not colloidally stable and showed rapid agglomeration and precipitation, therefore not all experiments were possible in the LB medium (see below).
Bacterial growth curves in the absence of silver in both media are shown in Fig. 1 and 2. Both bacterial strains showed a reduced growth in RPMI/10% FCS compared to the cultivation in LB. This effect was more pronounced for S. aureus compared to E. coli. The reason is that the LB medium is optimized for rapid bacterial growth, and therefore a bacterial growth rate reduction in RPMI/10% FCS which was originally developed for human cell culture is not unexpected.
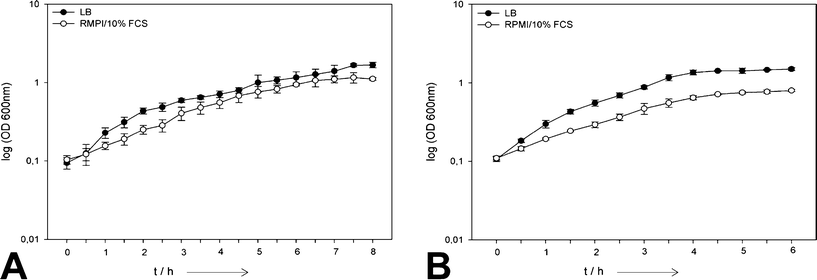 | ||
| Fig. 1 Comparison of the growth of planktonic E. coli (A) and S. aureus (B) cells in LB and RPMI/10% FCS in the absence of silver (N = 6). | ||
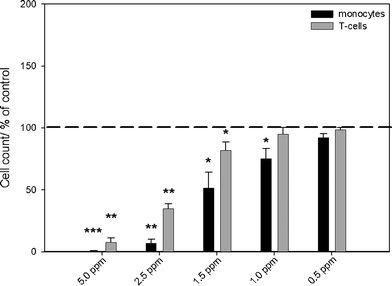 | ||
| Fig. 2 Effects of silver acetate on the viability of monocytes and T-cells, as analyzed by flow cytometry. The cells were treated with different concentrations of silver acetate for 24 h under cell culture conditions. Cell viability was determined by measuring the cell number by counting cell events for 30 s under constant flow. The data are expressed as the mean ± standard deviation (N = 3), given as the percentage of the control (cells cultured without silver, dashed line). Asterisks (*) indicate significant differences in comparison to the control (* p<0.05, ** p<0.01, *** p<0.001). | ||
The antibacterial effects of silver ions and silver nanoparticles on planktonic S. aureus and E. coli were studied by the determination of the minimum inhibitory concentrations (MICs, bacteriostatic) and minimum bactericidal concentrations (MBCs, bactericidal) both in LB and RPMI/10% FCS (Tables 1–4).
| Silver acetate (N = 4) | ||
|---|---|---|
| Cell density/mL−1 | MIC/ppm Ag | MBC/ppm Ag |
| 1 × 103 | 3.5 | 3.5 to 5 |
| 1 × 104 | 3.5 | 7.5 |
| 1 × 105 | 3.5 to 5 | 7.5 |
| 1 × 106 | 5 | 7.5 |
| Silver acetate (N = 9) | Silver nanoparticles (N = 6) | ||
|---|---|---|---|
| Cell density/mL−1 | MIC/ppm Ag | MBC/ppm Ag | MBC/ppm Ag |
| 1 × 103 | 0.5 to 1 | 0.5 to 1.25 | 12.5 to 20 |
| 1 × 104 | 0.75 | 1 to 1.25 | 20 |
| 1 × 105 | 1 to 1.25 | 2.5 | 50 |
| 1 × 106 | 1.25 to 2.5 | 2.5 to 5 | >50 |
| Silver acetate (N = 4) | ||
|---|---|---|
| Cell density/mL−1 | MIC/ppm Ag | MBC/ppm Ag |
| 1 × 103 | 2.5 | 5 |
| 1 × 104 | 3.5 | 7.5 |
| 1 × 105 | 3.5 | 7.5 |
| 1 × 106 | 5 | 7.5 |
| Silver acetate (N = 9) | Silver nanoparticles (N = 4) | ||
|---|---|---|---|
| Cell density/mL−1 | MIC/ppm Ag | MBC/ppm Ag | MBC/ppm Ag |
| 1 × 103 | 0.5 | 1.25 | 20 |
| 1 × 104 | 0.75 | 2.5 | 40 |
| 1 × 105 | 1.25 | 2.5 | >50 |
| 1 × 106 | 2.5 | 5 | >50 |
Silver showed a dose-dependent antimicrobial effect on both the tested strains. There was a slight difference in the susceptibility of gram-negative E. coli and gram-positive S. aureus to silver acetate. S. aureus seems to be less susceptible towards silver, suggesting that the about ten times thicker gram-positive cell wall with multiple murein layers and teichoic acid protects the cell against silver ions. A similar observation was reported by Amato et al.34 The antibacterial concentrations (MBCs) of the silver ions were higher in LB medium compared to those in RPMI/10% FCS, at least when smaller bacterial cell numbers were used for inoculation. This effect is probably due to the differences in the composition between both culture media. A decrease of bacterial cell growth was already observed within RPMI/10% FCS compared to the LB medium (Fig. 1), therefore a lower silver concentration may be already sufficient for bacterial growth inhibition and cell death in RPMI/10% FCS medium. However, several other factors may also contribute to this effect like a different complexation by proteins of the media and a different precipitation of poorly soluble salts like AgCl.29,35
Experiments with silver nanoparticles in LB medium were not possible because of the agglomeration of the particles. Thus, experiments on MICs and MBCs of silver nanoparticles were only performed in RPMI medium containing 10% (v/v) fetal calf serum, which stabilized the silver nanoparticles against agglomeration.15 The bactericidal concentrations (MBCs) of the silver nanoparticles were in a similar range compared to the literature data for silver nanoparticles of different sizes.36,37
We have previously shown that a given amount of silver ions is much more toxic to human cells than the same amount of silver in the form of silver nanoparticles.15,38,39 This was also observed here for the antibacterial thresholds of silver ions and nanoparticles. In general, the biological effect of silver is believed to be due to the free silver ion.2,40,41 Sotiriou and Pratsinis have argued, based on Ag/SiO2 nanoparticles from flame spray synthesis, that smaller silver particles release silver ions faster, leading to a higher toxicity due to a higher effective silver ion concentration. They found an antibacterial effect between 1 and 30 ppm silver concentration, depending on the silver nanoparticle size and the ratio of Ag![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SiO2.9
SiO2.9
Typically, the methods to determine the MIC and MBC are sensitive to variations in the number of bacteria. As shown in Table 1, an increase in the MIC and MBC of the silver preparations occurred when the inoculated cell number was increased. Comparable laboratory results have been described especially for β-lactam antibiotics.42 This phenomenon is known as the inoculum effect.43
Cell culture experiments using human blood and tissue cells were performed with human mesenchymal stem cells (hMSCs) and PMBCs (T-lymphocytes and monocytes). Both silver nanoparticles and silver ions showed toxic effects above a critical concentration (Fig. 2–5).
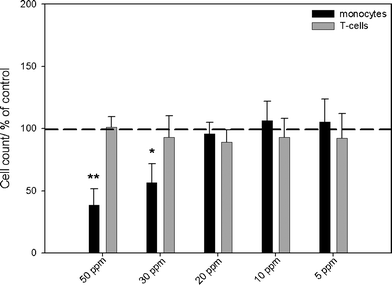 | ||
| Fig. 3 Effects of silver nanoparticles on the viability of monocytes and T-cells, as analyzed by flow cytometry. The cells were incubated in the presence of different concentrations of silver nanoparticles for 24 h under cell culture conditions. Cell viability was determined by measuring the cell number by counting cell events for 30 s under constant flow. The data are expressed as the mean ± standard deviation (N = 3), given as the percentage of the control (cells cultured without silver, dashed line). Asterisks (*) indicate significant differences in comparison to the control (* p<0.05, ** p<0.01). | ||
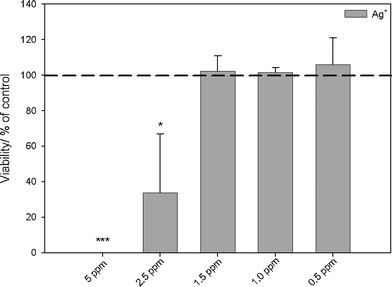 | ||
| Fig. 4 Influence of silver ions in the form of silver acetate on the viability of human mesenchymal stem cells (hMSCs). The cells were treated with different concentrations of silver acetate for 24 h under cell culture conditions. Vital cells (green fluorescence) were quantified by digital image processing (phase analysis). The data are expressed as mean ± standard deviation (N = 3) given as the percentage of the control (cells cultured without silver). Asterisks (*) indicate significant differences in comparison to the control (* p<0.05, *** p<0.001). | ||
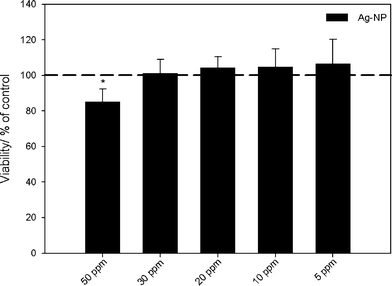 | ||
| Fig. 5 Influence of silver nanoparticles on the viability of human mesenchymal stem cells (hMSCs). The cells were treated with different concentrations of PVP-coated silver nanoparticles for 24 h under cell culture conditions. Vital cells (green fluorescence) were quantified by digital image processing (phase analysis). The data are expressed as mean ± standard deviation (N = 3) given as the percentage of the control (cells cultured without silver). An asterisk (*) indicates significant differences in comparison to the control (* p<0.05). | ||
When silver ions (silver acetate) were used, an increasing, dose-dependent cytotoxicity was determined in monocytes at concentrations above 1.0 ppm. However, T-cell viability was affected only at a silver concentration above 1.5 ppm (Fig. 2). After incubation of PBMCs with concentrations of silver nanoparticles of 30 ppm and higher, an increasing cytotoxicity towards monocytes was observed by flow cytometry (Fig. 3). No cytotoxic reactions were observed for T-cells after silver nanoparticle exposure under the experimental conditions. Previously, we have shown that the uptake rate of silver nanoparticles and the cell activation after exposure of silver is a cell type-dependent process. While agglomerates of silver nanoparticles were detected within the cytoplasm of peripheral monocytes, no agglomerated silver was found in the T-cells, as visualized by light microscopy, flow cytometry and focused ion beam/scanning electron microscopy (FIB/SEM).39 This may explain the different cytotoxicity observed in these two cell types. Control experiments with PVP-functionalized polystyrene nanoparticles showed no cytotoxic effects induced by the PVP itself (data not shown).
Human mesenchymal stem cells (hMSCs) as typical tissue-like cells were cultured in the presence of silver acetate or silver nanoparticles, and the viability of the treated cells was analyzed by fluorescence microscopy. The toxicity of silver (Fig. 4) and silver nanoparticles (Fig. 5) clearly increased with concentration, in accordance with the earlier results.13–16
All cytotoxicity data are comprised in Table 5. Remarkably, the critical concentrations were in the same range for bacteria and eukaryotic cells. To compare bacteria and human cells, it is more appropriate to use the MBC as this corresponds to the complete killing of the cells, rather than just the inhibition of cell division or proliferation.
| Silver acetate | Silver nanoparticles | |
|---|---|---|
| Escherichia coli | 0.5 to 2.5 (MIC) 0.5 to 5 (MBC) | n.p (MIC) 12.5 to >50 (MBC) |
| Staphylococcus aureus | 0.5 to 2.5 (MIC) 1.25 to 5 (MBC) | n.p (MIC) 20 to >50 (MBC) |
| T lymphocytes (T-cells) | 1.5 | >50 |
| Monocytes | 1 | 30 |
| Human mesenchymal stem cells (hMSCs) | 2.5 | 50 |
The cytotoxic concentrations for bacteria and human cells were in the same concentration range both for silver acetate and silver nanoparticles, respectively, with a factor of five at most between the lowest and highest concentration. Silver ions were more cytotoxic than silver nanoparticles by a factor of 20 to 30, but their toxic effects on bacteria and human cells occurred in the same concentration range.
A variety of silver-based dressings have become available which were approved for clinical use such as for the therapy of burn wounds.44 Remarkably, there is still no general agreement in the clinical efficacy of such treatments, even adverse effects such as a prolongation of healing time were reported.45 Brett has summarized the literature of silver toxicity towards bacteria and concluded that the typical bactericidal silver concentration is about 1 ppm, but that the presence of organic material can strongly increase this value.2 Albers et al. reported on the deleterious effects of different silver forms at antibacterial concentrations on osteoblasts and osteoclasts in vitro, supporting our results.46 Jain et al. have studied the effect of silver nanoparticles with 7–20 nm diameter on different bacterial strains and found critical concentrations of 0.78–6.25 ppm (MIC) and 12.5 ppm (MBC). For Hep G2 cells, they found an IC50 of 251 ppm.25 This agrees well with our results. In a comparative study with the reaction of S. epidermis and NIH 3T3 fibroblasts towards silver-impregnated polymer films, Agarwal et al. showed that these films can kill bacteria while cells remain unharmed.47 Vasilev et al. have studied the effect of silver nanoparticle-loaded plasma-polymerized films on S. epidermis and SaOs-2 osteoblasts and found a stronger effect towards bacteria than towards cells.26 D'Britto et al. have observed the same phenomenon with silver-releasing polymer scaffolds, three bacterial strains and CHO-K1 ovarian cells.28 Due to the basically unknown concentrations in the vicinity of such silver-releasing films, a strict comparison with our data is not possible, but the results point into the same direction.
Previous studies indicate that silver ions may precipitate as silver chloride or silver phosphate, that they may be complexed by proteins and that they may be possibly reduced.2,29 However, as the cell culture medium and the culture conditions were the same in the experiments presented here, the effects will be the same for bacteria and mammalian cells.
In general, it is important to note that the toxic effects on bacteria and mammalian cells are not strictly comparable due to the strongly different proliferation and growth rates. Whereas in bacteria, the inhibition of growth is typically followed easily by photometric approaches (cell density), in mammalian cells with much slower growth rates, the viability is considered. The different cell size and nature of the cell wall will definitely also play major roles. However, for a biomedical application as a bactericidal agent, it is of prime importance that the surrounding tissue (i.e. eukaryotic cells) is not harmed while bacterial cell growth is inhibited and bacterial cells are killed.
Conclusions
In general, the advantages of silver due to its bactericidal action must we weighed against possible tissue damage due to the cytotoxic nature of silver. In vitro cell culture has obvious limitations as a model for infected tissue sites, i.e. the in vivo situation with dynamic blood flow. However, our results clearly indicate a small effective window for silver in nanoparticulate or ionic form towards mammalian and bacterial cells. Cutting it short, silver is harming or killing bacteria in the same concentration range where it harms human cells (as present in surrounding tissue). This raises concerns about the widespread application of silver as an antibacterial agent and in medical applications and also in consumer commodities.Acknowledgements
We thank the Mercator foundation for support with a Mercur startup grant (to B.S., M.K., and M.E.). We thank the Deutsche Forschungsgemeinschaft for support within the priority program SPP 1313 BioNanoResponses (to M.E. and M.K.).References
- V. Alt, T. Bechert, P. Steinrücke, M. Wagener, P. Seidel, E. Dingeldein, E. Domann and R. Schnettler, Biomaterials, 2004, 25, 4383–4391 CrossRef CAS.
- D. W. Brett, Ostomy/wound manage, 2006, 52, 34–41 Search PubMed.
- S. W. P. Wijnhoven, W. J. G. M. Peijnenburg, C. A. Herberts, W. I. Hagens, A. G. Oomen, E. H. W. Heugens, B. Roszek, J. Bisschops, I. Gosens, D. van De Meent, S. Dekkers, W. H. De Jong, M. van Zijverden, A. J. A. M. Sips and R. E. Geertsma, Nanotoxicology, 2009, 3, 109–138 CrossRef CAS.
- B. Nowack, H. F. Krug and M. Height, Environ. Sci. Technol., 2011, 45, 1177–1183 CrossRef CAS.
- H. J. Johnston, G. Hutchison, F. M. Christensen, S. Peters, S. Hankin and V. Stone, Crit. Rev. Toxicol., 2011, 40, 328–346 CrossRef.
- J. W. Alexander, Surg. Infect., 2009, 10, 289–292 CrossRef.
- H. J. Klasen, Burns, 2000, 26, 131–138 CrossRef CAS.
- H. J. Klasen, Burns, 2000, 26, 117–130 CrossRef CAS.
- G. A. Sotiriou and S. E. Pratsinis, Environ. Sci. Technol., 2010, 44, 5649–5654 CrossRef CAS.
- K. N. J. Stevens, O. Crespo-Biel, E. E. M. van den Bosch, A. A. Dias, M. L. W. Knetsch, Y. B. J. Aldenhoff, F. H. van der Veen, J. G. Maessen, E. E. Stobberingh and L. H. Koole, Biomaterials, 2009, 30, 3682–3690 CrossRef CAS.
- D. Roe, B. Karandikar, N. Bonn-Savage, B. Gibbins and J. B. Roullet, J. Antimicrob. Chemother., 2008, 61, 869–876 CrossRef CAS.
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramirez and M. J. Yacaman, Nanotechnology, 2005, 16, 2346–2353 CrossRef CAS.
- L. Liu and R. H. Hurt, Environ. Sci. Technol., 2010, 44, 2169–2175 CrossRef.
- J. Liu, D. A. Sonshine, S. Shervani and R. H. Hurt, ACS Nano, 2010, 4, 6903–6913 CrossRef CAS.
- S. Kittler, C. Greulich, J. Diendorf, M. Köller and M. Epple, Chem. Mater., 2010, 22, 4548–4554 CrossRef CAS.
- C. M. Ho, S. K. W. Yau, C. N. Lok, M. H. So and C. M. Che, Chem.–Asian J., 2010, 5, 285–293 CrossRef CAS.
- W. Zhang, Y. Yao, N. Sullivan and Y. Chen, Environ. Sci. Technol., 2011, 45, 4422–4428 CrossRef CAS.
- L. W. Toy and L. Macera, Journal of the American Academy of Nurse Practitioners, 2011, 23, 183–192 CrossRef.
- K. Chaloupka, Y. Malam and A. M. Seifalian, Trends Biotechnol., 2010, 28, 580–588 CrossRef CAS.
- M. Ahamed, M. Karns, M. Goodson, J. Rowe, S. M. Hussain, J. J. Schlager and Y. Hong, Toxicol. Appl. Pharmacol., 2008, 233, 404–410 CrossRef CAS.
- R. M. Slawnson, H. Lee and J. T. Trevors, Biol. Met., 1990, 3, 151–154 CrossRef.
- J. P. Ruparelia, A. K. Chatterjee, S. P. Duttagupta and S. Mukherji, Acta Biomater., 2008, 4, 707–716 CrossRef CAS.
- E. M. Luther, Y. Koehler, J. Diendorf, M. Epple and R. Dringen, Nanotechnology, 2011, 22, 375101 CrossRef.
- C. Greulich, J. Diendorf, T. Simon, G. Eggeler, M. Epple and M. Köller, Acta Biomater., 2011, 7, 347–354 CrossRef CAS.
- J. Jain, S. Arora, J. M. Rajwade, P. Omray, S. Khandelwal and K. M. Paknikar, Mol. Pharmaceutics, 2009, 6, 1388–1401 CrossRef CAS.
- K. Vasilev, V. Sah, K. Anselme, C. Ndi, M. Mateescu, B. Dollmann, P. Martinek, H. Ys, L. Ploux and H. J. Griesser, Nano Lett., 2009, 10, 202–207 CrossRef.
- A. Agarwal, K. M. Guthrie, C. J. Czuprynski, M. J. Schurr, J. F. McAnulty, C. J. Murphy and N. L. Abbott, Adv. Funct. Mater., 2011, 21, 1863–1873 CrossRef CAS.
- V. D'Britto, H. Kapse, H. Babrekar, A. A. Prabhune, S. V. Bhoraskar, V. Premnath and B. L. V. Prasad, Nanoscale, 2011, 3, 2957–2963 RSC.
- S. Kittler, C. Greulich, J. S. Gebauer, J. Diendorf, L. Treuel, L. Ruiz, J. M. Gonzalez-Calbet, M. Vallet-Regi, R. Zellner, M. Köller and M. Epple, J. Mater. Chem., 2010, 20, 512–518 RSC.
- H. Wang, X. Qiao, J. Chen and S. Ding, Colloids Surf., A, 2005, 256, 111–115 CrossRef CAS.
- C. Greulich, J. Diendorf, J. Geβmann, T. Simon, T. Habijan, G. Eggeler, T. A. Schildhauer, M. Epple and M. Köller, Acta Biomater., 2011, 7, 3505–3514 CrossRef CAS.
- C. N. Lok, C. M. Ho, R. Chen, Q. Y. He, W. Y. Yu, H. Sun, P. K. H. Tam, J. F. Chiu and C. M. Che, JBIC, J. Biol. Inorg. Chem., 2007, 12, 527–534 CrossRef CAS.
- D. English and B. R. Andersen, J. Immunol. Methods, 1974, 5, 249–252 CrossRef CAS.
- E. Amato, Y. A. Diaz-Fernandez, A. Taglietti, P. Pallavicini, L. Pasotti, L. Cucca, C. Milanese, P. Grisoli, C. Dacarro, J. M. Fernandez-Hechavarria and V. Necchi, Langmuir, 2011, 27, 9165–9173 CAS.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. B. Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS.
- M. A. Ansari, H. M. Khan, A. A. Khan, A. Malik, A. Sultan, M. Shahid, F. Shujatullah and A. Azam, Biology and Medicine, 2011, 3, 141–146 CAS.
- G. A. Martinez-Castanon, N. Nino-Martinez, F. Martinez-Gutierrez, J. R. Martinez-Mendoza and F. Ruiz, J. Nanopart. Res., 2008, 10, 1343–1348 CrossRef CAS.
- S. Kittler, C. Greulich, M. Köller and M. Epple, Materialwiss. Werkstofftech., 2009, 40, 258–264 CrossRef CAS.
- C. Greulich, S. Kittler, M. Epple, G. Muhr and M. Köller, Langenbecks Arch. Surg., 2009, 394, 495–502 CrossRef CAS.
- Y. Matsumura, K. Yoshikata, S. I. Kunisaki and T. Tsuchido, Appl. Environ. Microbiol., 2003, 69, 4278–4281 CrossRef CAS.
- X. Chen and H. J. Schluesener, Toxicol. Lett., 2008, 176, 1–12 CrossRef CAS.
- I. Brook, Clin. Infect. Dis., 1989, 11, 361–368 CrossRef CAS.
- K. I. Udekwu, N. Parrish, P. Ankomah, F. Baquero and B. R. Levin, J. Antimicrob. Chemother., 2009, 63, 745–757 CrossRef CAS.
- I. Chopra, J. Antimicrob. Chemother., 2007, 60, 447–448 CrossRef CAS.
- M. N. Storm-Versloot, C. G. Vos, D. T. Ubbink and H. Vermeulen, Cochrane Database Syst. Rev., 2010, CD006478 Search PubMed.
- C. E. Albers, W. Hofstetter, K. A Siebenrock, R. Landmann and F. M. Klenke, Nanotoxicology, 2012 DOI:10.3109/17435390.2011.626538.
- A. Agarwal, T. L. Weis, M. J. Schurr, N. G. Faith, C. J. Czuprynski, J. F. McAnulty, C. J. Murphy and N. L. Abbott, Biomaterials, 2010, 31, 680–690 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2012 |
