Significant proteins affecting cerebral vasospasm using complementary ICPMS and MALDI-MS
Renee N.
Easter
ab,
Colin G.
Barry
b,
Gail
Pyne-Geithman
c and
Joseph A.
Caruso
*a
aDepartment of Chemistry, University of Cincinnati, Cincinnati, OH 45221, USA. E-mail: joseph.caruso@uc.edu
bForensic Chemistry Center, U.S. Food and Drug Administration, Cincinnati, Ohio 45237, USA
cDepartment of Neurosurgery, University of Cincinnati, Cincinnati, OH 45267, USA
First published on 6th October 2011
Abstract
Cerebral vasospasm (CV) following subarachnoid hemorrhagic stroke affects more than one million people each year. The etiology and prevention of CV is currently of great interest to researchers in various fields of medical science. More recently, the idea that selenium could be playing a major role in the onset of cerebral vasospasm has come into the spotlight. This study focused on using newly established metallomics techniques in order to explore the proteome associated with CV and if selenium might affect the discovered proteins. Size exclusion chromatography coupled to inductively coupled plasma mass spectrometry, along with LC-MALDI-TOF/TOF were both essential in determining protein identifications in three different sample types; a control (normal, healthy patient, CSF control), SAH stroke patients (no vasospasm, CSF C) and SAH CV patients (CSF V). The results of this study, although preliminary, indicate the current methods are applicable and warrant further application to these clinically important targets.
Introduction
Selenium is an essential micronutrient and plays an important role in many aspects of human health and disease.1 The main role of selenium in the body is to protect against oxidative stress. Selenocysteine (SeCys) is known as the 21st amino acid. When located with the selenocysteine insertion sequence (SECIS element) the UGA codon, typically a stop codon, leads to incorporation of selenocysteine into proteinsviaselenocysteine tRNA.2 To date there about 30 identified selenoproteins, 15 of which have been characterized.3 One major class of selenoproteins is peroxidases,3,4 which contain five Se-containing glutathione peroxidases (GPx). GPx1 is a selenium-dependent enzyme, which plays a major role in the antioxidant capacity of mammals.5 The selenocysteine residue (aa 49) is an essential component of the active site of GPx1. GPx1 is expressed in the cytoplasm of nearly all mammalian tissues.6GPx catalyzes the breakdown of hydrogen peroxide, a known reactive oxygen species (ROS). It has long been suspected that ROS play a major role in the etiology of cerebral vasospasm (CV) after subarachnoid hemorrhagic stroke (SAH) either by induction of the vasospasm itself7–11 or prevention of vessel relaxation.12,13 SAH, a type of hemorrhagic stroke, typically results from a ruptured aneurysm in the cerebrospinal fluid (CSF) compartment. In approximately 50% of patients surviving the initial hemorrhage, CV leads to intracranial vessel constriction (Fig. 1), typically in the vicinity of the hemorrhage, manifesting as delayed neurological deficit, this is a significant cause of mortality and morbidity in this patient population.
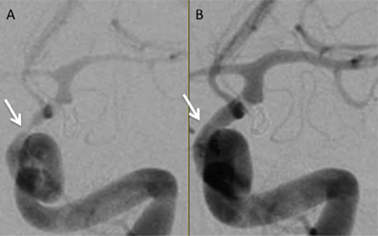 | ||
| Fig. 1 Left internal carotid artery angiogram in the arterial phase shown in a frontal projection. Both angiograms are from the same patient taken only minutes apart. Panel A shows severe spasm of the intracranial internal carotid artery (arrow) and middle cerebral artery prior to treatment. Panel B shows resolution of spasm after balloon angioplasty of the internal carotid artery and middle cerebral artery and infusion of verapamil (10 mg) into the internal carotid artery (courtesy Todd Abruzzo, MD, Mayfield Clinic). | ||
Worldwide, SAH and its sequela, CV, kill or seriously debilitate an estimated 1.2 million people annually of all ages, ethnic groups and gender. Recent studies found elevated levels of selenium and increased GPx activity in patients with SAH followed by CV.14,15 One of the barriers to research progress in prevention or reversal of CV after SAH is the ability to predict which SAH patients will develop vasospasm, and elucidation of the signaling pathways that contribute to cerebral vasospasm. This problem has been approached by phosphorylation studies,16,17 and in this study, selenium is utilized to study proteomic differences across the three sample types; namely CSF, from SAH patients (with or without CV).
Previous work by the Pyne-Geithman group14 has shown that the activity of GPx1 is significantly higher in SAH CSF from CV patients compared with non-CV SAH CSF. The amount of GPx1 protein was not different between groups, leading us to examine selenium levels. Selenium is an essential cofactor and integral part of the GPx enzyme. The Se(−), found in resting state, is first oxidized by the peroxide to SeOH which then reacts with a GSH molecule to form GS-Se and water, then by another GSH molecule to from Se(−) again, releasing GS-SG as the by-product. This mechanism of GS-SG formation depends on the selenium cofactor and thus the activity of GPx is affected as well.
Earlier studies from this laboratory have shown that proteomic analysis of cerebral spinal fluid yields useful and potentially important information in terms of differentiation between SAH patients who do, or do not, progress into cerebral vasospasm.16,17 Taken in toto these studies are likely to provide important insights into identifying yet undiscovered biomarkers that could potentially be used to aid in diagnosis, theranostics, and treatment decision-making in this and other patient populations. Further studies will require statistically acceptable numbers of patient samples across the three groups.
In this study, both elemental (ICPMS) and molecular (MALDI-TOF/TOF) identification are utilized for exploring whether or not selenium is associated with CV relevant proteins that may be found in cerebral spinal fluid samples. The study covers a control (normal, healthy patient, CSF control) sample, SAH stroke patient samples (no vasospasm, CSF C) and SAH CV patient samples(CSF V). SEC-ICPMS was used to allow for the identification of selenium peaks and then MALDI-TOF/TOF was employed for protein identification after using the peaks screened for Se.
Experimental
Materials
Tris-HCl and alpha-cyano-4-hydroxycinnamic acid (CHCA) were purchased from Thermo Scientific (Rockford IL, USA). Ammonium bicarbonate as well as analytical grade methanol, acetonitrile, and glacial acetic acid were purchased from Fisher Scientific (Springfield NJ, USA). 10x Tris/Tricine/SDS buffer, Coomassie Brilliant blue R-250 stain, 4x XT sample buffer and criterion XT 4–12% bis-tris precast gels were purchased from Bio-Rad (Hercules CA, USA). Dithiothreitol (DTT) was purchased from Pierce Chemicals (Rockford IL, USA). Iodoacetamide (IAA) was purchased from Sigma Aldrich (St. Louis MO, USA) and trypsin was purchased from Promega (Madison WI, USA).Cerebrospinal fluid
Five freeze dried cerebral spinal fluid samples were received from the Department of Neurosurgery at the University of Cincinnati. CSF was obtained from patients admitted to the NSICU with appropriate local and Federal IRB compliance. The samples were assayed for total protein using the BCA protein assay.18 Samples were classified as control (no SAH), CSF C (SAH, no vasospasm) or CSF V (SAH and vasospasm). Control CSF was obtained under an IRB exemption, from outpatient clinics where normal CSF is drained for pseudotumor cerebri or non-obstructive hydrocephalus therapy. CSF C and CSF V patients received identical standard of care in the Neurosciences Intensive Care Unit at the University Hospital, Cincinnati. All patients had intraventricular drains in place, and received nutrition via a nasogastric tube. Patients all received FiberSource HN (a complete tube feeding nutritional liquid, Nestle Nutrition, Highland Park, MI) during the period when CSF was collected. This supplement contains sodium selenite equivalent to 17.5 μg per meal, and patients receive 3 meals a day.Instrumentation
Chromatographic analysis was performed on an Agilent 1100 series HPLC system (Santa Clara, CA, USA) equipped with a binary pump; a vacuum chambered micro degasser, and a thermostatically controlled column compartment. Isotopes of selenium were monitored using an Agilent 7500cx ICPMS (Santa Clara, CA, USA) equipped with an on-axis rf only octopole collision reaction cell. All data analysis was performed using Agilent chromatographic software package. Molecular mass spectrometry was performed on an AB Sciex 5800 MALDI-TOF/TOF (Foster City, CA, USA) equipped with an on-axis 1KHz laser. All data analysis was performed through ProteinPilotTM software 3.0. Gel electrophoresis was performed using Criterion gels with a Power Pac 200 power source from Bio-Rad (Hercules, CA, USA).Size exclusion Chromatography-ICPMS
Samples were reconstituted in 1 mL of 18 MΩ water, to a final concentration of 30 mg of protein/mL. Corresponding sample types were pooled to give three different samples; control (CSF N), subarachnoid hemorrhagic stroke patients (CSF C) and subarachnoid hemorrhagic stroke patients with vasospasm (CSF V). Samples were then separated isocratically (0.400 mL min−1) in 30 mM Tris-HCl buffer, pH 7.5 on a Superdex 200 size exclusion column. Selenium isotopes of m/z 77, 78 and 82 were monitored on the ICPMS using hydrogen in the collision reaction cell. Instrumental parameters can be found in Table 1. The column was calibrated with a protein mix that included thyroglobulin (670 kDa), IGG (150 kDa), albumin (67 kDa), myoglobin (44 kDa) and vitamin B12 (1.35 kDa).| Instrument/parameters | Settings |
|---|---|
| HPLC parameters | |
| Column flow rate | 0.400 mL min−1 |
| Mobile phase A | 30 mM Tris-HCl, pH 7.5 |
| Isocratic separation | 0–65 min, 100% M.P. A |
| ICPMS parameters | |
| Spray chamber | Scott double pass total consumption spray chamber |
| Spray chamber temperature | 2 °C |
| Interface cones | Ni sample cone, Ni skimmer cone |
| Plasma power | 1500 W |
| RF matching | 1.7 V |
| Plasma gas flow | 1.02 L min−1 |
| Collision reaction cell gas | H2, 4.6 mL min−1 |
| Quadrupole bias | −15 V |
| Octopole bias | −18 V |
1D Gel electrophoresis
Selenium fractions were collected offline from the SEC-ICPMS experiment and further separated by 1D gel electrophoresis. 35 μL of each sample was mixed with 12.5 μL of sample buffer and 2.5 μL of XT Reducing Agent. The sample solutions were incubated for 5 min at 95 °C, allowed to cool to room temperature, then loaded onto a Criterion 4–12% bis tris precast gel. The running buffer used was a 20x dilution of XT MOPS buffer. A voltage of 200 V was applied to the gel for 1 h to achieve a separation. The gel was then washed 3x for ten minutes with deionized water, then stained with Coomassie Brilliant blue stain for 1 h. The gel was then destained with a solution containing 10% MeOH, 5% Acetic Acid and 85% water until background coloring was removed. Protein bands were cut out of the gel and were washed and digested using standard methods described below.In-gel protein digestion
Individual gel bands were cut from each lane of the gel for analysis. The fractions were dehydrated by covering the gel pieces with acetonitrile, the samples were then vortexed and the liquid removed via aspiration. The gel pieces were then re-swelled by the addition of 0.1 M ammonium bicarbonate (≈200 μL), the samples vortexed, followed by the removal of liquid by aspiration. These steps were repeated three times. 100 μL of ammonium bicarbonate was added to the fractions and the samples were then heated to 37 °C for 15 min. The liquid was aspirated and 100 μL of ACN was added and again heated to 37 °C for 15 min. The liquid was then removed by aspiration. Following the final dehydration, the gel pieces were opaque and colorless. 95 μL of ammonium bicarbonate was added to the fractions along with 10 μL of DTT for a final concentration of 10 mM DTT. The fractions were then incubated for 30 min at 37 °C. Following the incubation, the liquid was removed and 7.5 μL of IAA (15 mM final concentration) and 42.5 μL of ammonium bicarbonate were added to the fractions. The fractions were incubated in the dark for 30 min at room temperature. After 30 min, the liquid was removed by aspiration and 8 μL of trypsin, 5.0 × 105 ng mL−1 was added, along with enough ammonium bicarbonate to cover the fractions, and heated overnight at 37 °C. After overnight digestion, the liquid was transferred to new vials and saved. 200 μL of 50% aqueous ACN was added to the gel fractions. The fractions were sonicated for 20 min, then spun at 1500 × g for 10 min and the liquid removed and combined with the digestion solution from corresponding sample.MALDI-TOF/TOF
Digested proteins from the gel were spotted, using the dried droplet technique, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with CHCA (10 mg mL−1 in 70% aqueous ACN + 0.1% TFA) on a stainless steel MALDI target. Samples were analyzed in reflector positive mode and MS/MS was performed on the 50 most intense peaks with a S/N ≥ 20. Autotryptic fragments were excluded from the MS/MS search.
1 with CHCA (10 mg mL−1 in 70% aqueous ACN + 0.1% TFA) on a stainless steel MALDI target. Samples were analyzed in reflector positive mode and MS/MS was performed on the 50 most intense peaks with a S/N ≥ 20. Autotryptic fragments were excluded from the MS/MS search.
ProteinPilot software
MALDI-TOF/TOF data was analyzed through ProteinPilot™ software version 3.0, Applied Biosystems & MDS Inc. The raw data were searched against a human refseq database containing 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 668 entries using the Paragon™ algorithm version 3.0.0.0, 113
668 entries using the Paragon™ algorithm version 3.0.0.0, 113![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 442 and through Mascot against the same human refseq database. A trypsin enzyme was used in the search parameters with a maximum of 2 missed cleavages. Variable modifications that were included in the parameters were methionine oxidation and a cysteine carbamidomethylation.
442 and through Mascot against the same human refseq database. A trypsin enzyme was used in the search parameters with a maximum of 2 missed cleavages. Variable modifications that were included in the parameters were methionine oxidation and a cysteine carbamidomethylation.
Results and discussion
SEC-ICPMS
CSF samples were separated under isocratic conditions (100% mobile phase A for 65 min) with 30 mM Tris buffer at a pH of 7.5 at ambient temperature. All three samples were run in duplicate to ensure minimum reproducibility. Isotopes of Se were monitored in order to separate those peaks containing selenium. The chromatogram showed only three Se containing peaks (Fig. 2). These peaks eluted around the same time as the albumin, myoglobin and vitamin B12 standards. The first peak at 30-34 min was common to all three sample types. The second peak at 34-39 min was only present in CSF C and CSF V sample types. The third peak, which is in the exclusion volume for the column (∼45 min), was again common to all samples but there are differences within that peak. Since size exclusion chromatography separates based on size (hydrodynamic radii), it is possible that those peaks collected contained multiple proteins or peptides. As a result another dimension of separation was still needed. Fractions were collected offline for further separation and molecular identification.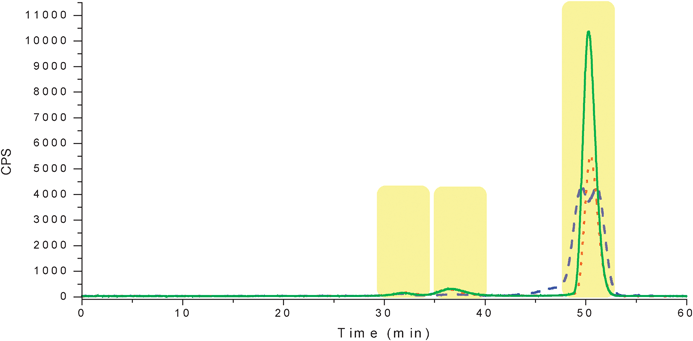 | ||
| Fig. 2 SEC-ICPMS chromatogram for selenium found in CSF samples. The blue dashed trace is the CSF control, the red dotted trace is CSF C and the green solid trace is CSF V. The highlighted portions are the fractions that were collected offline for further analysis. | ||
1DGel electrophoresis
Nine selenium fractions (three from each sample type) were further separated using 1-D gel electrophoresis. 35 μL of each fraction was mixed with 2.5 μL of reducing agent in order to break disulfide bonds. 12.5 μL of XT sample buffer was added to the sample. 40 μL of each sample was loaded into separate lanes on the gel, and a molecular weight ladder (BlueRanger prestained protein molecular weight marker mix, 16.8–220 kDa, Peirce, Rockford Il, USA) was placed on each end. The gel was run for 1 h at a constant 200 V. Each fraction from the SEC-ICPMS experiments resulted in multiple protein bands as shown in Fig. 3. The bands appeared to be in roughly the same molecular weight range as expected from the SEC experiments. Fraction 3 from each sample did not contain any protein bands. This was the most intense selenium peak and the last one to elute from the column, suggesting lower MW selenium. Fraction three was rerun on a 16.5% Tris/Tricine gel in order to determine if smaller proteins or peptides were present in the fraction. No stained bands were visualized on the peptide gel (data not shown). Fraction three was also investigated with electrospray ionization linear trap quadrupole mass spectrometry (ESI-LTQ-MS). No m/z corresponding to known selenium species such as selenomethionine or selenocysteine were detected. The protein bands from the gel then were cut into fractions and an in-gel digestion was performed to extract the proteins from the gel fractions.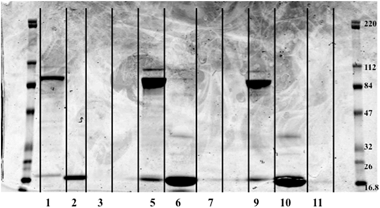 | ||
| Fig. 3 Image of 1-D gel. Image was obtained with an Odyssey Infra-red Imaging System from LI-COR, Lincoln Ne., USA. Far lanes contain molecular weight ladders while sample was loaded in the middle lanes. Lane 1 – CSF control Fraction #1, L2 – CSF control #2, L3 – CSF control #3, L5 – CSF C#1, L6 – CSF C#2, L7 – CSF C #3, L9 – CSF V #1, L10 – CSF V #2, L11 – CSF V #3, Lanes 4, 8, 12 – Blank. Fraction 3 for each sample had no stained protein bands present. Molecular weight of standards is in kDa. | ||
MALDI analysis of gel fractions
After digestion, each fraction was spotted (≈0.75 μL) on a stainless steel MALDI target. CHCA matrix (10 mg mL−1) was spotted on top of each sample spot in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The samples were analyzed in reflector positive mode with MS/MS being performed on the 50 most intense peaks from each spectrum (Fig. 4 and 5). The data were then analyzed using ProteinPilot software and 6 proteins were identified (Table 2). For each protein identified, the peptide mapping was performed by the software as well, in order to verify the peptides that were used for the protein identification (Table 3). Additionally manual peptide mapping was performed in order to verify the software results. Two of the proteins were albumin and hemoglobin, which were virtual certainties. The other proteins, although usually found in high abundance, were of some interest and are further discussed below.
1 ratio. The samples were analyzed in reflector positive mode with MS/MS being performed on the 50 most intense peaks from each spectrum (Fig. 4 and 5). The data were then analyzed using ProteinPilot software and 6 proteins were identified (Table 2). For each protein identified, the peptide mapping was performed by the software as well, in order to verify the peptides that were used for the protein identification (Table 3). Additionally manual peptide mapping was performed in order to verify the software results. Two of the proteins were albumin and hemoglobin, which were virtual certainties. The other proteins, although usually found in high abundance, were of some interest and are further discussed below.
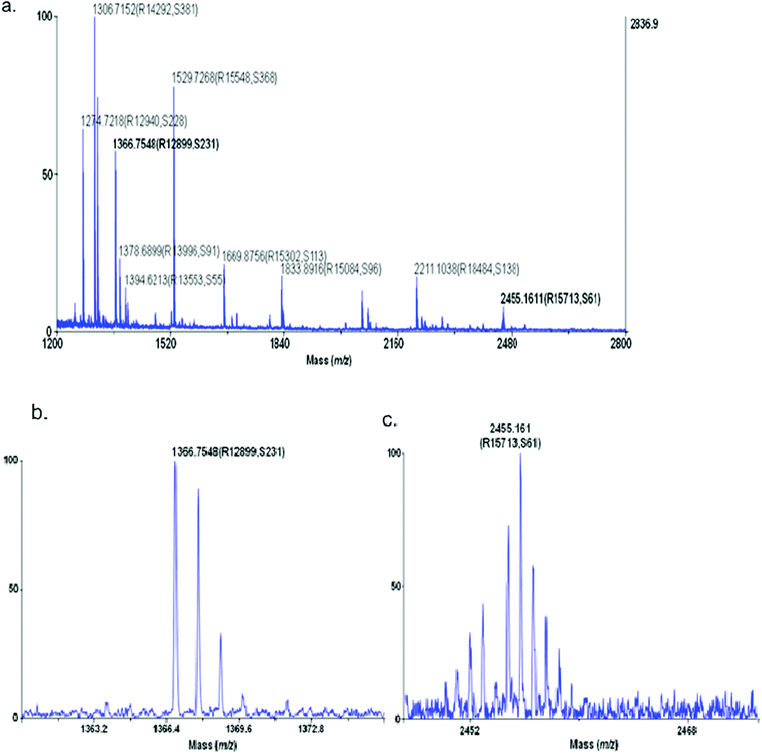 | ||
| Fig. 4 Precursor ion spectra of CSF V sample Fraction 1 (a) with zoomed detail for precursor ion m/z 1366.75 (b) and m/z = 2455.16 (c). The values in parenthesis are the resolution (R) and the signal to noise (S) determined by the software. | ||
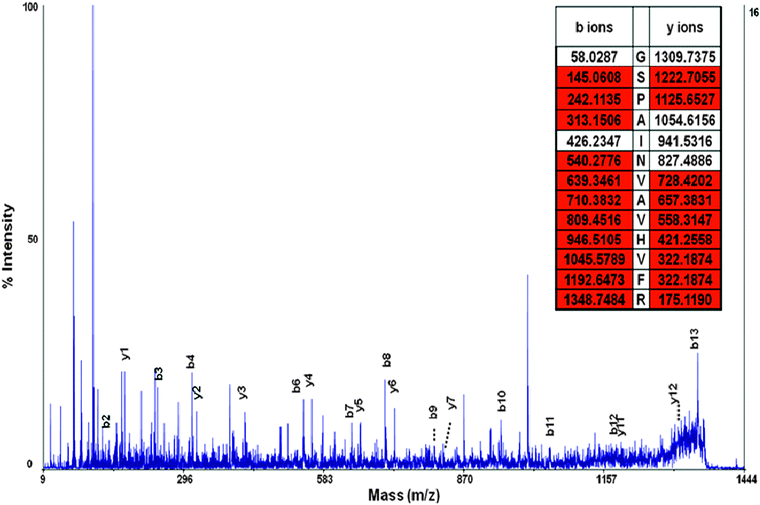 | ||
| Fig. 5 MS/MS spectra of peptide GSPAINVAVHVFR. y and b ions are labeled and the peptide has excellent sequence coverage. The peptide fragment was used to identify the transthyretin precursor protein, a protein known to be associated with selenium and CSF. | ||
| Protein | Unuseda | %Covb | %Cov (95%)c | Accession | Name | Species | Sample |
|---|---|---|---|---|---|---|---|
| a Unused column is equivalent to the score of other databases such as Mascot or Spectrummill. A score of 1.3 corresponds to a 95% confidence. A score of 2 or higher is a confident match and considered to be a good score for ProteinPilot. b % Coverage is the sequence coverage of the protein identified. c % Coverage (95%) is the sequence coverage pertaining to only high confidence peptides (95% or greater confidence). | |||||||
| 1 | 14.77 | 22.33 | 10.84 | gi|4502027 | Albumin preproprotein | Homo sapiens | All |
| 2 | 14.39 | 82.31 | 56.46 | gi|4504349 | Beta globin | Homo sapiens | All |
| 3 | 14 | 42.15 | 42.15 | gi|4502517 | Carbonic anhydrase I | Homo sapiens | C and V |
| 4 | 8.68 | 49.66 | 33.33 | gi|4507725 | Transthyretin precursor | Homo sapiens | Control and V |
| 5 | 2 | 5.53 | 5.53 | gi|4505591 | Peroxiredoxin 1 | Homo sapiens | V |
| 6 | 1.4 | 13.48 | 12.06 | gi|4503109 | Cystatin S precursor | Homo sapiens | C |
| Protein name(s) | Confidence | Sequence | Δ Mass | Precursor MW | Theoretical MW |
|---|---|---|---|---|---|
| Transthyretin precursor | 99 | AADDTWEPFASGK | 0.0231 | 1393.638 | 1393.615 |
| Transthyretin precursor | 99 | GSPAINVAVHVFR | 0.033242 | 1365.785 | 1365.752 |
| Transthyretin precursor | 99 | KAADDTWEPFASGK | 0.034216 | 1521.744 | 1521.71 |
| Transthyretin precursor | 99 | TSESGELHGLTTEEEFVEGIYK | 0.091569 | 2454.236 | 2454.144 |
| Transthyretin precursor | 72 | YTIAALLSPYSYSTTAVVTNPKE | 0.077644 | 2488.352 | 2488.2474 |
Protein discussion
Transthyretin precursorTransthyretin precursor is a 55-kDa homotetramer with a dimer of dimers configuration that is synthesized in the liver, choroid plexus and retinal pigment epithelium. Each monomer is a 127-residue polypeptide rich in beta sheet structure. It is typically associated with plasma and cerebral spinal fluid and its function is to transport thyroxine (T (4)) and retinol.19,20
Current studies have shown that an increase in transthyretin precursor can be an indicator of good clinical outcome from a cerebral infarction patient.21,22 Benvenga and coworkers studied the protein concentration of thirty different patients that had been hospitalized after a cerebral infarction. From their study, it was determined that the level of transthyretin was the most sensitive factor for recovery. Patients with decreasing transthyretin concentrations died, while an increase in concentration was found in all survivors.22
Transtyretin has also recently been shown to be upregulated by selenium supplementation. Sinhaand coworkers supplemented a group of men with selenium-enriched yeast to understand the effects of supplementation on serum protein expression. Nine months after supplementation, serum levels had shown that eight proteins were upregulated, including transthyretin. All of the proteins upregulated were redox sensitive proteins.23 From these studies, we can conclude that transthyretin is responsive to selenium concentrations and it shows differently between the stroke and the CV patients, thus making it the most interesting protein found in this study and one that needs further investigation for its possible CV biomarker role.
Cystatin X precursor
Cystatin X precursor, cystatin 3 or Cystatin C is a 120 amino acid protein that is commonly found in all tissues and bodily fluids. It is a potent inhibitor of lysosomal proteinases and probably one of the most important extracellular inhibitors of cysteine proteinases. Cystatin C belongs to the type 2 cystatin gene family.24–28Cystatin C was first described as ‘gamma-trace’ in 1961 as a trace protein in the CSF and urine of patients with renal failure.25 Since then, Cystatin 3 is routinely measured as a marker of glomerular filtration rate.27,29 More recently, genomic and proteomic studies consistently report that this precursor of Cystatin C is strongly associated with cerebral amyloid angiopathy,30,31 which is a known cause of intracerebral hemorrhage; deposition of amyloid in the vessel walls weakens them and facilitates aneurysm growth and ultimately rupture.32
Several studies have also found that increased levels of Cystatin C are associated with the risk of death, several types of cardiovascular disease (including myocardial infarction, stroke, heart failure, peripheral arterial disease and metabolic syndrome) and healthy aging.24,26,28,33–36 Of particular relevance to the patient group herein, Cystatin C levels are decreased in atherosclerotic (so-called ‘hardening’ of the arteries) and aneurysmal (saccular bulging) lesions of the aorta.36–38Breakdown of parts of the vessel wall in these conditions is thought to result from an imbalance between proteinases (cysteine proteases and matrix metalloproteinases, increased) and their inhibitors (such as cystatin C, decreased).34,36,38–40
Peroxiredoxin
Peroxiredoxins are a ubiquitous family of antioxidant enzymes that control cytokine-induced peroxide levels. In this way, they mediate signal transduction in mammalian cells.41 Peroxiredoxins are frequently referred to as alkyl hydroperoxide reductase (AhpC) in bacteria. Other names include thiol specific antioxidant (TSA). This family contains AhpC and TSA, as well as related proteins.41There are three classes of peroxiredoxins (Prxs): typical 2-Cys Prxs; atypical 2-Cys Prxs; and 1-Cys Prxs. These enzymes share the same basic catalytic mechanism, in which a redox-active cysteine (the peroxidatic cysteine) in the active site is oxidized to a sulfenic acid by the peroxide substrate. The recycling of the sulfenic acid back to a thiol is what distinguishes the three enzyme classes,42 2-Cys peroxiredoxins are reduced by thiols such as glutathione, while the 1-Cys enzymes may be reduced by ascorbic acid or glutathione in the presence of GST-π. Inactivation of these enzymes by over-oxidation of the active thiol to sulfinic acid can be reversed by sulfiredoxin.43–46
Recent evidence has revealed that post-translational modifications of peroxiredoxin impair function and contribute to pathology.43,47,48 The sulfur present in the active site (Cys) can exchange with selenium, which could sufficiently alter the peroxiredoxin and could possibly lead to a change in oxidative stress. It is known that oxidative stress is a contributing factor in a number of chronic neurodegenerative pathologies as well as acute cerebrovascular disorders such as stroke. It is becoming clear that the thioredoxin-peroxiredoxin system represents an increasingly attractive therapeutic target for central nervous system disorders associated with oxidative stress. These findings suggest more detailed studies need to be done.
Carbonic anhydrase
Carbonic anhydrases (CAH) belong to a family of enzymes that catalyze the rapid conversion of carbon dioxide and water to bicarbonate and protons. Most CAHs contain a zinc ion at the active site; they are classified as metalloenzymes.49 The primary function of the enzyme in animals is to interconvert carbon dioxide and bicarbonate to maintain acid–base balance in blood and other tissues,50 and to help transport carbon dioxide out of tissues.51CAH has been the focus of pharmacological therapeutics for a number of years. Carbonic anhydrase inhibitors are a class of pharmaceuticals that suppress the activity of CAH. Their clinical use has been established as antiglaucoma agents, diuretics, antiepileptics, in the management of mountain sickness, gastric and duodenal ulcers, neurological disorders, and osteoporosis.52CAH expression increases after ischemic stroke,53 and has been postulated as a CSF marker for cerebrovascular disease.54 SAH followed by CV in its manifestation, generates a blockage that mimics an ischemic stroke.
Conclusions
Of the proteins identified, all may be feasible as biomarkers of CV and SAH, but they must be studied with statistically viable sample numbers. And further, with increased sample numbers, it will be possible to search for lower copy numbers of regulatory proteins. The most promising protein found appears to be the transthyretin precursor and to study in more detail, the selenium association.Size exclusion chromatography coupled with inductively coupled plasma mass spectrometry has proven to be a useful metallomics tool for the search of proteins in biological samples when used in tandem with molecular mass spectrometry. Although only high abundance proteins have been identified, some likely play a critical role in cerebral vasospasm. A comparison of the three pools of sample types is tentative, but with the methods established, we are now moving to a larger sample set for full verification of the comparisons made. Multidimensional fractionation techniques currently under investigation will allow for the identification of lower abundance proteins where the selenium containing peptides/proteins more likely exist at fewer copy numbers, and the role that these play in the debilitating CV disease.
Acknowledgements
The authors wish to thank Agilent Technologies for instrumentation support and the US FDA STEP program for financial support for RNE. We would also like to thank Todd Abruzzo, MD, Mayfield Clinic, for Fig. 1.References
- M. P. Rayman, Lancet, 2000, 356, 233–241 CrossRef CAS.
- R. Walczak, E. Westhof, P. Carbon and A. Krol, RNA, 1996, 2, 367–379 CAS.
- K. Brown and J. R. Arthur, Public Health Nutr., 2001, 4, 593–599 CrossRef CAS.
- J. Lu and A. Holmgren, J. Biol. Chem., 2009, 284, 723–727 CrossRef CAS.
- U. Schweizer, A. U. Bräuer, J. Köhrle, R. Nitsch and N. E. Savaskan, Brain Res. Rev., 2004, 45, 164–178 CrossRef CAS.
- S. Comhair and S. C. Erzurum, Antioxid. Redox Signaling, 2005, 7, 72–79 CrossRef CAS.
- F. M. Faraci, J. Appl. Physiol., 2006, 739–743 CrossRef CAS.
- F. R. Sharp, J. Cereb. Blood Flow Metab., 2006, 10, 1223–1233 Search PubMed.
- K. Osuka, Y. Watanabe, N. Usuda, K. Atsuzawa, T. Wakabayashi and M. Takaysu, Brain Res., 2010, 1332, 12–19 CrossRef CAS.
- G. J. Pyne-Geithman, C. J. Morgan, K. Wagner, E. M. Dulaney, J. Carrozzella, D. S. Kanter, M. Zuccarello and J. F. Clark, Journal Of Cerebral Blood Flow And Metabolism: Official Journal Of The International Society Of Cerebral Blood Flow And Metabolism, 2005, 25, 1070–1077 CrossRef CAS.
- J. F. Clark, M. Reilly and F. R. Sharp, J. Cereb. Blood Flow Metab., 2002, 22, 472–478 CrossRef CAS.
- K. Irani, Circ. Res., 2000, 87, 179–183 CAS.
- R. Motterlini, R. Foresti, M. Intaglietta and R. M. Winslow, Am. J. Physiol., 1996, 270(Heart Circ. Physiol.), H107–H114 CAS.
- G. J. C. Pyne-Geithman, N. Danielle, P. Prakash and J. F. Clark, Neurol. Res., 2009, 31, 195–199 CrossRef CAS.
- J. M. Siverling, University of Cincinnati, Cincinnati, 2009.
- J. Ellis, E. Del Castillo, M. Montes Bayon, R. Grimm, J. F. Clark, G. J. Pyne-Geithman, S. Wilbur and J. A. Caruso, J. Proteome Res., 2008, 7, 3747–3754 CrossRef CAS.
- K. Kroening, J. Kuhlmann, R. Easter, J. F. Clark, G. J. Pyne-Geithman and J. A. Caruso, Metallomics, 2010, 2, 334–341 RSC.
- P. K. Smith, et al. , Anal. Biochem., 1985, 150(1), 76–85 CrossRef CAS.
- M. A. Liz, F. M. Mar, F. Franqunho and M. M. Sousa, IUBMB Life, 2010, 62, 429–435 CAS.
- C. E. Fleming, F. M. Mar, F. Franqunho and M. M. Sousa, Int. Rev. Neurobiol., 2009, 87, 337–346 CrossRef CAS.
- C. Gao, B. Zhang, W. Zhang, S. Pu, J. Yin and Q. Gao, Clin. Exp. Med., 2010 Search PubMed.
- S. Benvenga, L. Morgante, L. Bartalena, L. Manna, L. Li Calzi, M. A. Coraci and F. Trimarchi, Ann. Clin. Res., 1986, 18, 203–207 CAS.
- R. Sinha, L. Sinha, N. Facompre, S. Russell, R. I. Somiari, J. P. J. Ritchie and K. El-Bayoumy, Cancer Epidemiol., Biomarkers Prev., 2010, 19, 2332–2340 CrossRef CAS.
- W. Koenig, D. Twardella, H. Brenner and D. Rothenbacher, Clin. Chem., 2005, 51, 321–327 CAS.
- H. Löfberg and A. O. Grubb, Scand. J. Clin. Lab. Invest., 1979, 39, 619–626 CrossRef.
- A. Servais, P. Giral, M. Bernard, E. Bruckert, G. Deray and I. Bagnis, Am. J. Med., 2008, 121, 426–432 CrossRef CAS.
- O. Simonsen, A. Grubb and H. Thysell, Scand. J. Clin. Lab. Invest., 1985, 45, 97–101 CrossRef CAS.
- B. Zethelius, L. Berglund, J. Sundström, E. Ingelsson, S. Basu, A. Larsson, P. Venge and J. Arnlöv, N. Engl. J. Med., 2008, 358, 2107–2116 CrossRef CAS.
- A. Grubb, O. Simonsen, G. Sturfelt, L. Truedsson and H. Thysell, Acta Med. Scand., 1985, 218, 499–503 CrossRef CAS.
- S. A. Kaeser, M. C. Herzig, J. Coomaraswamy, E. Kilger, M. L. Selenica, D. T. Winkler, M. L. Staufenbiel, A. Grubb and M. Jucker, Nat. Genet., 2007, 39, 1437–1439 CrossRef CAS.
- E. Levy, M. Jaskolski and A. Grubb, Brain Pathol., 2006, 16, 60–70 CrossRef CAS.
- A. Pezzini, E. Del Zotto, I. Volonghi, A. Giossi, P. Costa and A. Padovani, Curr. Med. Chem., 2009, 16, 2498–2513 CrossRef CAS.
- P. Eriksson, K. G. Jones, L. C. Brown, R. M. Greenhalgh, A. Hamsten and J. T. Powell, Br. J. Surg., 2004, 91, 86–89 CrossRef CAS.
- M. Gacko, L. Chyczewski and L. Chrostek, Pol. J. Pathol., 1999, 50, 83–86 CAS.
- E. Levy, C. Lopez-Otin, J. Ghiso, D. Geltner and B. Frangione, J. Exp. Med., 1989, 169, 1771–1778 CrossRef CAS.
- G.-P. Shi, G. K. Sukhova, A. Grubb, A. Ducharme, L. H. Rhode, R. T. Lee, P. M. Ridker, P. Libby and H. A. Chapman, J. Clin. Invest., 1999, 104, 1191–1197 CrossRef CAS.
- H. Abdul-Hussein, R. G. V. Soekhoe, E. Weber, K. H. von der Thüsen, R. Kleeman, A. Mulder, J. H. van Bockel, R. Hanemaaijer and J. H. N. Lindeman, Am. J. Pathol., 2007, 170, 809–817 CrossRef.
- J. S. Lindholdt, E. J. Erlandson and E. W. Henneberg, Br. J. Surg., 2001, 88, 1472–1475 CrossRef.
- S. Abisi, K. G. Burnand, M. Waltham, J. Humphries, P. R. Taylor and A. J. Smith, J. Vasc. Surg., 2007, 46, 1260–1266 CrossRef.
- E. Bengtsson, J. Nilsson and S. Jovinge, Front. Biosci., 2008, 13, 5780–5786 CrossRef CAS.
- H. Chae and S. Rhee, Biofactors, 1994, 4, 177–180 CAS.
- T. J. Jonsson and W. T. Lowther, Subcell. Biochem., 2007, 44, 115–141 CrossRef.
- G. Montiero, B. B. Horta, D. C. Pimenta, O. Augusto and L. E. Netto, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 4886–4891 CrossRef.
- L. Poole, Arch. Biochem. Biophys., 2005, 433, 240–254 CrossRef CAS.
- S. Rhee, H. Chae and K. Kim, Free Radical Biol. Med., 2005, 38, 1543–1552 CrossRef CAS.
- Z. Wood, E. Scroder, J. Robin Harris and L. Poole, Trends Biochem. Sci., 2003, 28, 32–40 CrossRef CAS.
- K. F. Bell and G. E. Hardingham, Antiox. Redox Signal., 2010 Search PubMed.
- L. Flohé, Methods Enzymol., 2010, 473, 1–39 Search PubMed.
- S. Lindskog, Pharmacol. Ther., 1997, 74, 1–20 CrossRef CAS.
- V. Lyall, R. I. Alam, D. Q. Phan, G. L. Ereso, T. H. Phan, S. A. Malik, M. H. Montrose, S. Chu, G. L. Heck, G. M. Feldman and J. A. DeSimone, Am. J. Physiol. (Cell Physiol), 2001, 281, C1005–1013 CAS.
- S. Breton, JOP, 2001, 2, 159–164 CAS.
- C. T. Suparan and A. Scozzafava, Bioorg. Med. Chem., 2007, 15, 4336–4350 CrossRef.
- Y. Tang, H. Xu, L. Lit, W. Walker, A. Lu, R. Ran, J. P. Gregg, M. Reilly, A. Pancioli, J. C. Khoury, L. R. Sauerbeck, J. A. Carrozzella, J. Spilker, J. F. Clark, K. R. Wagner, E. C. Jauch, D. J. Chang, P. Verro, J. P. Broderick and F. R. Sharp, J. Cereb. Blood Flow Metab., 2006, 26, 1089–1102 CrossRef CAS.
- A. K. Parkkila, S. Parkkila, M. Reunanen, O. Niemela, S. Tuisku, I. Rautakorpi and H. Rajaniemi, Eur. J. Clin. Invest., 1997, 27, 392–397 CAS.
| This journal is © The Royal Society of Chemistry 2012 |
