Click JAHAs: conformationally restricted ferrocene-based histone deacetylase inhibitors†‡
John
Spencer
*a,
Jahangir
Amin
a,
Ramesh
Boddiboyena
a,
Graham
Packham
b,
Breeze E.
Cavell
b,
Sharifah S.
Syed Alwi
b,
Ronald M.
Paranal
c,
Tom D.
Heightman§
d,
Minghua
Wang
d,
Brian
Marsden
d,
Peter
Coxhead
e,
Matthew
Guille
e,
Graham J.
Tizzard
f,
Simon J.
Coles
f and
James E.
Bradner.
c
aSchool of Science, University of Greenwich at Medway, Chatham, ME4 4TB, UK. E-mail: j.spencer@gre.ac.uk; Fax: +44-2083319805; Tel: +44-2083318215
bCancer Research UK Centre, University of Southampton, Faculty of Medicine, Mailpoint 824, Southampton General Hospital, Tremona Road, Southampton, SO16 6YD, UK
cDana-Farber Cancer Institute, 44 Binney Street, Dana Building, D510D, Boston, MA 02115, USA
dStructural Genomics Consortium, University of Oxford, Old Road Campus Research Building, Old Road Campus, Roosevelt Drive, Headington, Oxford, OX3 7DQ, UK
eSchool of Biological Sciences, King Henry Building, University of Portsmouth, Portsmouth, PO1 2DT, UK
fUK National Crystallography Service, School of Chemistry, University of Southampton, Highfield, Southampton, SO17 1BJ, UK
First published on 20th October 2011
Abstract
A small library of ferrocene-based 1,2,3-triazole-containing hydroxamic acids has been synthesised employing click chemistry: 7-(4-ferrocenyl-1H-1,2,3-triazol-1-yl)-N-hydroxyheptanamide, 4b, containing the 1,2,3-triazole moiety adjacent to the ferrocene group, displayed excellent HDAC inhibition and activity in cells, inhibiting the deacetylation of tubulin as well as inducing cell cycle arrest.
Introduction
Epigenetic regulation of gene expression plays a key role in the aetiology of cancer and other diseases.1 Increased levels of histone deacetylases (HDACs) are associated with transcriptional silencing and cancer progression by inducing a condensed chromatin structure.2 The use of HDAC inhibitors (HDACis) can result in open, hyperacetylated chromatin, resulting in restored transcription of tumour suppressor genes, apoptosis and cell cycle arrest.3A number of HDACis have been described and a common pharmacophore has been established, namely an aryl cap combined with a linker and a zinc binding motif; the latter is often a hydroxamic acid or benzamide group.4 The HDACi SAHA has been successfully developed for the treatment of cutaneous T-cell lymphoma.5 Attempts to conformationally restrict the long chain alkyl linker in SAHA, via “click” chemistry, in order to reduce entropy upon enzyme binding, modify isoform selectivity and enhance its pharmacokinetics have been reported (Fig. 1).6,7
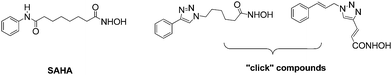 | ||
| Fig. 1 SAHA and conformationally restricted HDACis. | ||
Synthesis
Having recently identified JAHAs (parent JAHA is shown, N1-(ferrocenyl)-N8-hydroxyoctanediamide, Fig. 1) as potent ferrocene-based HDACis,8 we now disclose the synthesis and biological investigation of triazole-based analogues, namely click JAHAs, since they may offer the potential to act as metal-based probes for cocrystal structure determinations of HDACs. We have adopted two distinct synthetic strategies for the synthesis of the click products. The first (Scheme 1) involves the introduction of the triazole unit adjacent to the ferrocene “cap” as in 4. Hence, the cycloaddition of an azide1 with ferroceneacetylene2 was followed by conversion of the ester3 into a hydroxamic acid. The X-ray structure of the intermediate 3b was determined and established the presence of the triazole moiety as well as its expected 1,4-regiochemistry.9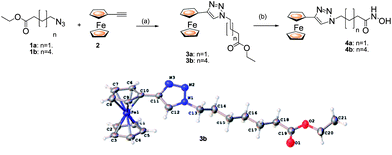 | ||
| Scheme 1 Reagents and conditions: (a) CuI (20 mol %), DIPEA (1.1 eq.), 60 °C, 1 h. (b) NH2OH·HCl, KOH. | ||
A second approach involved placing the heterocyclic group adjacent to the hydroxamic acid zinc binding group as in 7 and 10 (Scheme 2). Here, a ferrocene-based azide5 or 8 was combined with an alkyne followed by treatment with hydroxylamine. We assume this regiochemistry from the well established 1,4-regiochemistry with Cu catalysis and by comparison with the X-ray data for 3b.9
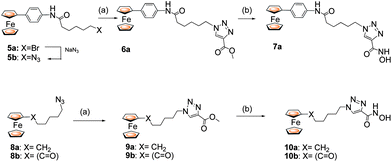 | ||
| Scheme 2 Reagents and conditions: (a) CuI (20 mol %), DIPEA (1.1 eq.), 60 °C, 1 h. (b) NH2OH·HCl, KOH. | ||
The cocrystal structure of SAHA in HDAC8 has been determined and provides a useful model for docking studies of HDACis.10 Hence, docking of 4b predicted it to have good HDAC inhibitory action against HDAC8 (Fig. 2 (a)). However, the vicinity of the triazole unit to the hydroxamic acid in 10a was assumed to be detrimental to HDAC8 binding due to steric reasons (Fig. 2(b)). SAHA and 11 were previously docked into HDAC8.8
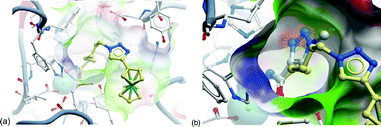 | ||
| Fig. 2 Docking analysis of click JAHAs in HDAC8 (based on PDB code: 1t69). (a) 4b docked in HDAC8; hydroxamate can readily bind to zinc. (b) Overlay of 10a over 4b (ferrocene groups omitted for clarity). A triazole adjacent to the hydroxamic acid group in 10a leads to a steric clash with the protein: (using Glide from Schrodinger for docking and ICM from Molsoft for preparation of the Figure). | ||
Biological activity
Biological evaluation of the click JAHAs has been performed as described in our previously disclosed work (Table 1).8,11 As can be seen, analogue 4b displays excellent HDAC inhibition with mainly single digit nM IC50 values, which are slightly lower than that of SAHA (albeit, our compounds have higher MWs compared to SAHA). The shorter chained derivative 4a exhibited weaker inhibition, highlighting the importance of chain length for HDAC inhibition, albeit its relatively low IC50 value towards HDAC6 is promising (69 nM). Compounds 7a and 10a were devoid of HDAC inhibition, consistent with previous findings that chain branching close to the hydroxamic acid moiety is not tolerated sterically by HDACs4 and predicted by docking studies on 10a for HDAC8. The related complex 10b, however, displayed single digit micromolar IC50 towards HDACs 1–3 and 8 and a submicromolar IC50 towards HDAC6. Compound 4a represents a useful lead in the design of isoform-selective tool compounds to probe tubulin acetylation. As found with other JAHAs, poor to moderate inhibition of HDAC8 was observed for all click JAHAs although 4b had an IC50 = 183 nM. Consistent with other JAHAs, no significant inhibition of Class IIa HDACs (4,5,7,9) was observed (data not shown).8| IC50 (μM) | HDAC1 | HDAC2 | HDAC3 | HDAC6 | HDAC8 |
|---|---|---|---|---|---|
| a IC50 values for compounds (μM) with SD in parentheses. Assays were performed in duplicate. b — denotes no appreciable activity. | |||||
| 4a | 1.2765 (0.1137) | 2.6357 (0.2476) | 0.7856 (0.0860) | 0.0693 (0.0044) | 1.5312 (0.1200) |
| 4b | 0.0045 (0.0003) | 0.0095 (0.0007) | 0.0072 (0.0004) | 0.0026 (0.0002) | 0.1834 (0.0081) |
| 7a | — | — | — | — | — |
| 10a | — | — | — | — | — |
| 10b | 1.5520 (0.1654) | 3.0107 (0.4598) | 1.4128 (0.1985) | 0.2948 (0.0583) | 2.4190 (0.1436) |
| 11 | 0.0792 (0.0042) | 0.1711 (0.0105) | 0.0842 (0.0051) | 0.0100 (0.0008) | 0.0411 (0.0021) |
| SAHA | 0.0206 (0.0013) | 0.0466 (0.0044) | 0.0198 (0.0016) | 0.0033 (0.0005) | 0.6875 (0.0628) |
We next wished to demonstrate cell-based activity for our click JAHAs. Hence, the latter were evaluated for growth inhibition in breast cancer cells (MCF7). From a preliminary screen at 10 μM concentration, only 4b showed appreciable growth inhibition, hinting again at the poor cellular penetration of a number of these compounds.8 Compound 4b was re-evaluated and registered an IC50 (±sd) = 575 ± 154 nM (n = 2), comparable to SAHA (730 ± 380 nM).8
Given that only 4b displayed any appreciable HDAC inhibition and cytotoxicity, we anticipated that it would have sufficient cellular penetration for flow cytometry experiments. Propidium iodide-staining experiments demonstrated that 4b induced cell cycle arrest and cell death in MCF7 cells (Table 2). Consistent with effects of other JAHAs, 4b decreased the proportion of cells in the G1 and S phases of the cell cycle, associated with an accumulation of cells in G2/M. Similarly, 4b increased the proportion of cells with sub-G1 DNA content, a marker of cell death, compared with a DMSO control.
|
|
|||||
|---|---|---|---|---|---|
| Effect of 4b on cell cycle phases | Effect of 4b on cell death | ||||
| G1 | S | G2/M | sub-G1 | ||
| a 1 μM concentration, 48h, MCF7 cells. Mean of two experiments ± range. | |||||
| DMSO | 59 ± 2 | 11 ± 1 | 31 ± 3 | DMSO | 4 ± 2 |
| 4b | 48 ± 2 | 5 ± 1 | 45 ± 2 | 4b | 45 ± 11 |
Compound 4b was slightly less effective than either SAHA or the analogue 11 at promoting tubulin (HDAC6 substrate) and lysine acetylation (in chromatin) by flow cytometry, possibly due to reduced cellular permeability due to the presence of the ferrocene and triazole units (Fig. 3).4,8
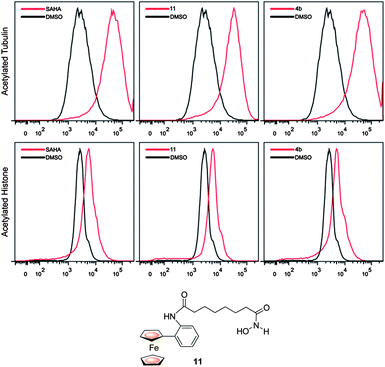 | ||
| Fig. 3 Comparative flow cytometry cell-based data for 4b, 11 and SAHA. | ||
To confirm HDAC6 inhibition in vivo, Xenopus laevis embryos were treated with 4b or 11 at concentrations varying between 10 μM and 200 μM.12,13,14HDAC6 activity in the embryos was monitored by assessing the acetylation levels of alpha tubulin. Both compounds blocked deacetylation with compound 11 being effective at 10 μM and compound 4b at 100 μM, similar to SAHA (Fig. 4). The discrepancy could be due to the reduced cellular penetration of the click analogues, as indicated above, although we cannot rule out a speciation effect i.e. a differential activity of the inhibitors on human HDAC6 (the enzyme used for biochemical profiling) and the XenopusHDAC6 homologue (76% human vs. Xenopus homology).
 | ||
| Fig. 4 Sets of 12 2-cell Xenopus laevis embryos were treated with compound 4b (a) compound 11 (b) or SAHA (c) at the concentrations shown until they developed to stage 14. Protein extract equivalent to one embryo was separated by duplicate SDS-PAGE gels and acetylated alpha-tubulin or total alpha-tubulin detected by Western blotting. | ||
Conclusion
A series of conformationally restricted JAHAs have been synthesised and one analogue 4b was shown to have good HDAC inhibition in both biochemical ex vivo and in vivo assays. JAHAs and, to a lesser extent, due to their limited activities and cellular penetration, click JAHAs hold great potential as metal-based probes for elucidating the role of HDACs in cancer and in other areas of biochemical interest.Acknowledgements
Greenwich University (GRE and the Alumni fund), BP are thanked for financial assistance (J.S. and J.A., for equipment), CRUK (G.P.), the Malaysian Government (S.S.S.A.), the Biochemical and Biophysical Sciences Research Council, University of Southampton and the Watercress Alliance (B.E.C.). J.E.B. and R.M.P. are supported by grants from the Multiple Myeloma Research Foundation and the National Institutes of Health. S.J.C. and G.J.T. would like to thank the EPSRC for funding. The EPSRC National Mass Spectrometry Service Centre (Swansea University) is thanked for HRMS measurements. José de Jesús Cázares Marinero, ENSCP (Paris), is thanked for helping rename our ferrocene compounds. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.Notes and references
- J. M. Annemieke, A. J. M. De Ruijter, A. H. Van Gennip, H. N. Caron, S. Kemp and A. Van Kuilenburg, Biochem. J., 2003, 370, 737–749 CrossRef CAS.
- J. D. Best and N. Carey, Drug Discovery Today, 2010, 15, 65–70 CrossRef CAS.
- P. A. Marks, Expert Opin. Invest. Drugs, 2010, 19, 1049–1066 CrossRef CAS.
- M. Paris, M. Porcelloni, M. Binaschi and D. Fattori, J. Med. Chem., 2008, 51, 1505–1529 CrossRef CAS.
- P. A. Marks and R. Breslow, Nat. Biotechnol., 2007, 25, 84–90 CrossRef CAS.
- J. Shen, R. Woodward, J. P. Kedenburg, X. Liu, M. Chen, L. Fang, D. Sun and P. G. Wang, J. Med. Chem., 2008, 51, 7417–7427 CrossRef CAS.
- P. C. Chen, V. Patil, W. Guerrant, P. Green and A. K. Oyelere, Bioorg. Med. Chem., 2008, 16, 4839–4853 CrossRef CAS.
- The IC50 values for SAHA, 11 and 12 are somewhat higher than those previously reported, due to a different batch of enzyme and technique employed, but the trends in terms of excellent HDAC6 and HDAC8 inhibition remain: J. Spencer, J. Amin, M. Wang, G. Packham, S.S.S. Alwi, G.J. Tizzard, S.J. Coles, R.M. Paranal, J.E. Bradner and T.D. Heightman, ACS Med. Chem. Lett., 2011, 2, 358–362 Search PubMed.
- H. C. Kolb and K. B. Sharpless, Drug Discovery Today, 2003, 8, 1128–1137 CrossRef CAS.
- J. R. Somoza, R. J. Skene, B. A. Katz, C. Mol, J. D. Ho, A. J. Jennings, C. Luong, A. Arvai, J. J. Buggy, E. Chi, J. Tang, B. C. Sang, E. Verner, R. Wynands, E. M. Leahy, D. R. Dougan, G. Snell, M. Navre, M. W. Knuth, R. V. Swanson, D. E. McRee and L. W. Tari, Structure, 2004, 12, 1325–1334 CrossRef CAS.
- J. E. Bradner, N. West, M. L. Grachan, E. F. Greenberg, S. J. Haggarty, T. Warnow and R. Mazitschek, Nat. Chem. Biol., 2010, 6, 238–243 CrossRef CAS.
- M. Guille. Molecular Methods in Developmental Biology: Xenopus and Zebrafish, Totowa, N.J.: Humana Press, 1999 Search PubMed.
- C. Robinson and M. Guille, Mol. Methods Dev. Biol., 1999, 127, 89–97 Search PubMed.
- J. Spencer, J. Amin, S. K. Callear, G. J. Tizzard, S. J. Coles, P. Coxhead and M. Guille, Metallomics, 2011, 3, 600–608 RSC . Example: Synthesis of 4b: (i) Synthesis of 7-azido-heptanoic acid ethyl ester, 1b. In an oven dried round bottom flask (100 mL) equipped with a reflux condenser, stirrer and under an inert atmosphere, ethyl-7-bromoheptanoate (1.20 cm3, 5.00 mmol) and sodium azide (0.65 g, 10.00 mmol) were dissolved in reagent grade DMF (2.5 cm3) and warmed at 90 °C for 1.5 h. After cooling, the reaction mixture was quenched with deionised water (20 cm3) and extracted with ethyl acetate (20 cm3). The organic layer was washed with sat. brine (3 × 20 cm3), separated then dried over magnesium sulphate. Concentration of the volatiles gave 1b (0.55 g, 59%) which was used in the next step without any further purification. 1H NMR (δH, 300.0 MHz, CDCl3) 1.23 (3H, J 6.0 Hz, CH3), 1.30–1.43 (4H, m, 2xCH2), 1.51–1.66 (4H, m, 2xCH2), 2.27 (2H, t, J 9.0 Hz, CH2), 3.24 (2H, t, J 6.0 Hz, CH2), 4.10 (2H, q, J 9.0 Hz, CH2); 13C NMR (δ, 75.0 MHz, CDCl3); 14.1, 24.7, 26.3, 28.5, 34.1, 51.3, 60.1, 60.3, 171.1. (ii) Synthesis of ferrocenyl 1,4-triazole-heptanoic acid ethyl ester, 3b. 1b (0.18 g, 1.00 mmol), ferrocenyl ethylene 2 (0.23 g, 1.10 mmol) and copper iodide (38.1 mg, 0.20 mmol) were combined in N,N-diisopropylethylamine (0.19 cm3, 1.10 mmol), t-BuOH (1 cm3) and deionised water (1 cm3). The reaction mixture was warmed to 60 °C for 1 h. The cooled reaction mixture was extracted using ethyl acetate (10 cm3) and washed with aqueous ammonium chloride (20 cm3) and sat. brine (20 cm3). The organic layer was dried over magnesium sulphate before being filtered then concentrated. The organic layer was initially filtered through a 2 cm pad of Celite before being purified by silica gel column chromatography using 3
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 ethyl acetate/hexane to give the product 3b (0.34 g, 85%) as a dark orange solid which was recrystallised from 2
7 ethyl acetate/hexane to give the product 3b (0.34 g, 85%) as a dark orange solid which was recrystallised from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ethyl acetate/hexane. M.p. 150–152 °C; 1H NMR (δH, 300.0 MHz, CDCl3) 1.25 (3H, t, J 6.0 Hz, CH3), 1.32–1.45 (4H, m, 2xCH2), 1.59–1.69 (2H, m, CH2), 1.90–1.97 (2H, m, CH2), 2.30 (2H, t, J 9.0 Hz, CH2), 4.07 (5H, s, C5H5), 4.12 (2H, m, CH2), 4.29 (2H, t, J 3.0 Hz, Fc), 4.35 (2H, t, J 9.0 Hz, CH2), 4.72 (2H, t, J 3.0 Hz, Fc), 7.45 (1H, s, CH); 13C NMR (δ, 67.0 MHz, CDCl3); 14.3, 24.6, 26.1, 28.4, 30.0, 34.0, 50.0, 60.2, 66.6, 68.6, 69.5, 75.7, 118.9, 146.5, 173.5; HRMS (m/z, HNESP) [M + H]+ for C21H28N3O2Fe = Calc. 408.1572 observed. 408.1570. Elemental anal. For C21H27FeN3O2 Calc. C, 61.6%; H, 6.6%, N, 10.3% observed C, 61.3%, H, 6.8%, N, 10.4%. (iii) Synthesis of 7-(4-ferrocenyl-1H-1,2,3-triazol-1-yl)-N-hydroxyheptanamide, 4b. Hydroxylamine hydrochloride (3.50 g, 50.00 mmol) was dissolved in reagent grade methanol (38 cm3) and potassium hydroxide (3.00 g, 50.00 mmol) was added slowly to this mixture at r.t. before it was warmed to 40 °C for approximately 20 min. After this time has elapsed the reaction mixture was cooled to 0 °C. The precipitate was filtered and the filtrate was added to 3b (0.10 g, 0.25 mmol), potassium hydroxide (0.30 g, 5.35 mmol) and stirred at r.t. overnight before being extracted with ethyl acetate (10 cm3) and washed with ammonium chloride (10 cm3), sat. brine (10 cm3), dried over magnesium sulphate. After filtration then solvent concentration, the product 4b was precipitated from 1
5 ethyl acetate/hexane. M.p. 150–152 °C; 1H NMR (δH, 300.0 MHz, CDCl3) 1.25 (3H, t, J 6.0 Hz, CH3), 1.32–1.45 (4H, m, 2xCH2), 1.59–1.69 (2H, m, CH2), 1.90–1.97 (2H, m, CH2), 2.30 (2H, t, J 9.0 Hz, CH2), 4.07 (5H, s, C5H5), 4.12 (2H, m, CH2), 4.29 (2H, t, J 3.0 Hz, Fc), 4.35 (2H, t, J 9.0 Hz, CH2), 4.72 (2H, t, J 3.0 Hz, Fc), 7.45 (1H, s, CH); 13C NMR (δ, 67.0 MHz, CDCl3); 14.3, 24.6, 26.1, 28.4, 30.0, 34.0, 50.0, 60.2, 66.6, 68.6, 69.5, 75.7, 118.9, 146.5, 173.5; HRMS (m/z, HNESP) [M + H]+ for C21H28N3O2Fe = Calc. 408.1572 observed. 408.1570. Elemental anal. For C21H27FeN3O2 Calc. C, 61.6%; H, 6.6%, N, 10.3% observed C, 61.3%, H, 6.8%, N, 10.4%. (iii) Synthesis of 7-(4-ferrocenyl-1H-1,2,3-triazol-1-yl)-N-hydroxyheptanamide, 4b. Hydroxylamine hydrochloride (3.50 g, 50.00 mmol) was dissolved in reagent grade methanol (38 cm3) and potassium hydroxide (3.00 g, 50.00 mmol) was added slowly to this mixture at r.t. before it was warmed to 40 °C for approximately 20 min. After this time has elapsed the reaction mixture was cooled to 0 °C. The precipitate was filtered and the filtrate was added to 3b (0.10 g, 0.25 mmol), potassium hydroxide (0.30 g, 5.35 mmol) and stirred at r.t. overnight before being extracted with ethyl acetate (10 cm3) and washed with ammonium chloride (10 cm3), sat. brine (10 cm3), dried over magnesium sulphate. After filtration then solvent concentration, the product 4b was precipitated from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ethyl acetate/hexane then filtered (0.047 g, 47%) as a light orange solid. M.p. 162–164 °C. 1H NMR (δH 500.0 MHz, DMSO-d6); 1.18–1.31 (4H, m, 2xCH2), 1.44–1.48 (4H, m, 2xCH2), 1.91 (2H, t, J 10.0 Hz, 4.01 (5H, s, C5H5), 4.28 (2H, brs, Fc), 4.32 (2H, t, J 5.0 Hz), 4.69 (2H, brs, Fc), 8.16 (1H, s, CH), 8.63 (1H, s, OH), 10.31 (1H, s, NH); 13C NMR (δC 100.5 MHz, CD3OD); 26.5, 27.1, 29.3, 31.0, 33.6, 51.2, 67.6, 69.8, 70.5, 76.0, 121.4, 147.9, 172.8; HRMS (m/z, HNESP) [M + H]+ for C19H25N4O2Fe = Calc. 395.1368 observed. 395.1365; elemental anal. For C19H24FeN4O2.12 moles of DCM (recrystallised from 2
20 ethyl acetate/hexane then filtered (0.047 g, 47%) as a light orange solid. M.p. 162–164 °C. 1H NMR (δH 500.0 MHz, DMSO-d6); 1.18–1.31 (4H, m, 2xCH2), 1.44–1.48 (4H, m, 2xCH2), 1.91 (2H, t, J 10.0 Hz, 4.01 (5H, s, C5H5), 4.28 (2H, brs, Fc), 4.32 (2H, t, J 5.0 Hz), 4.69 (2H, brs, Fc), 8.16 (1H, s, CH), 8.63 (1H, s, OH), 10.31 (1H, s, NH); 13C NMR (δC 100.5 MHz, CD3OD); 26.5, 27.1, 29.3, 31.0, 33.6, 51.2, 67.6, 69.8, 70.5, 76.0, 121.4, 147.9, 172.8; HRMS (m/z, HNESP) [M + H]+ for C19H25N4O2Fe = Calc. 395.1368 observed. 395.1365; elemental anal. For C19H24FeN4O2.12 moles of DCM (recrystallised from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 DCM/hexane); Calc. C, 56.5%, H, 6.0%, N, 13.8%; observed C, 56.5%, H, 5.9%, N, 13.4%. HPLC (anal.); (99
8 DCM/hexane); Calc. C, 56.5%, H, 6.0%, N, 13.8%; observed C, 56.5%, H, 5.9%, N, 13.4%. HPLC (anal.); (99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hexane/MeOH, 1 mL min−1 Rt = 26 min 2 s). % Purity = 95%.
1 hexane/MeOH, 1 mL min−1 Rt = 26 min 2 s). % Purity = 95%.
Footnotes |
| † This article is part of a MedChemComm web theme issue on epigenetics. |
| ‡ Electronic supplementary information (ESI) available: Biological procedures, synthesis of click JAHAs and X-ray crystallography. CCDC reference numbers 838910. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c1md00203a |
| § Current address: Astex Pharmaceuticals, 436 Cambridge Science Park, Cambridge, CB4 0QA, UK. |
| This journal is © The Royal Society of Chemistry 2012 |

