Targeting epigenetic modifiers: Inhibitors of histone methyltransferases
Elisabeth-Maria
Bissinger
a,
Ralf
Heinke
b,
Wolfgang
Sippl
b and
Manfred
Jung
*a
aInstitute of Pharmaceutical Sciences, Albert-Ludwigs-Universität Freiburg, Albertstr. 25, 79104, Freiburg, Germany. E-mail: manfred.jung@pharmazie.uni-freiburg.de
bDepartment of Pharmaceutical Chemistry, Martin-Luther University of Halle-Wittenberg, 06120, Halle/Saale, Germany
First published on 9th July 2010
Abstract
The term epigenetics is defined as inheritable changes that influence the outcome of a phenotype without changes in the genome. Epigenetics is based upon DNA methylation and posttranslational histone modifications. While there is much known about reversible acetylation as a posttranslational modification, research on histone methylation is still emerging, especially with regard to drug discovery. As aberrant epigenetic modifications have been linked to many diseases, inhibitors of histone methylation are very much in demand. This article gives an outline on the different histone methyltransferases, their involvement in disease, the available inhibitors and their potential as drugs.
 Elisabeth-Maria Bissinger | Elisabeth studied Pharmacy at the University of Münster. After graduation, she started in 2007 as a PhD student in the group of Manfred Jung at the Institute of Pharmaceutical Sciences in Freiburg. Her work focuses on the synthesis and in vitro testing of new histone methyltransferase inhibitors. |
 Ralf Heinke | Ralf studied Pharmacy at the Martin-Luther-University of Halle-Wittenberg and is a PhD student there in the group of Wolfgang Sippl at the Institute of Pharmacy since 2007. His work focuses on the computer-based design of histone methyltransferase inhibitors. |
 Wolfgang Sippl | Wolfgang studied Pharmacy at the University in Berlin. He obtained a PhD in Pharmaceutical Chemistry at the University of Düsseldorf in the group of Hans-Dieter Höltje and was a post-doctoral fellow at the Université Louis-Pasteur in Strasbourg (France) where he worked with Camille G. Wermuth. Wolfgang then took a senior researcher position in Düsseldorf before moving to the University of Halle-Wittenberg as a full professor for Medicinal Chemistry in 2003. Since 2010 he is Director of the Institute of Pharmacy in Halle. His main interests are focused on computational chemistry and structure-based drug design. |
 Manfred Jung | Manfred studied Pharmacy at the University of Marburg where he also did his PhD in the group of Prof. W. Hanefeld working on synthesis of aromatic retinoids. As a postdoctoral fellow he joined Prof. T. Durst (Ottawa, Canada) working on the synthesis of amino acid derivatives. From 1994 to 2003 he was a group leader at the University of Münster. Since 2003 Manfred is a professor of Pharmaceutical Chemistry at the University of Freiburg. His research focus is on Chemical Epigenetics, covering both inhibitor synthesis and in-vitro assay development. He has published over 80 papers, mainly on epigenetics. |
1. Introduction
The phenotype of an organism is based on its genetic information stored in the DNA but the final outcome of a certain phenotype is governed by transcriptional regulation. Epigenetics is defined as inheritable changes of the phenotype that take place without any changes in the genetic code. Key biochemical processes that define epigenetics are DNA methylation and histone modifications. In order to be packaged efficiently, DNA is wrapped around basic proteins, the histones. DNA and one histone octamer together form a nucleosome and the assembly of all nucleosomes is called chromatin. The N-termini of the histones are rich in arginine and lysine residues and protrude from the nucleosomes. They are subject to many types of post-translational modifications such as acetylation, methylation, phosphorylation, sumoylation and ubiquitinylation.1 These modifications can occur in different combinations at specific sites which is referred to as the “Histone Code” and explains why a great variety of gene expression patterns and thus different phenotypes is possible from one genetic code.2 In this article, we focus on histone methylation because DNA methylation and the role of other histone modifications have recently been reviewed more broadly in other publications.3–62. Protein methylation and biological significance
2.1 Histone methylation
Generally, histone methylation is involved in heterochromatin formation and maintenance, X-chromosome inactivation, transcriptional regulation, DNA repair, RNA maturation and genomic imprinting.7 Histones can be methylated either on lysine or on arginine residues. In contrast to lysines that can be mono-, di- or trimethylated, arginines are only mono- or dimethylated, but the dimethylation can be symmetrical or asymmetrical (see Fig. 1).8 Depending on the site of methylation, lysinemethylation can be linked either to transcriptional activation or repression respectively. Methylation on H3K4, H3K36 and H3K79 for example is considered to be activating whereas methylated H3K9, H3K27 and H4K20 are regarded to be repressive marks.8 Further, loss of trimethylation at H4K20 is a general hallmark of human cancer together with the loss of acetylation at H4K16.9 Just recently, Manuyakorn and co-workers have discovered that cellular histone modification patterns help to predict the prognosis of pancreatic adenocarcinoma. Low cellular levels of H3K4me2, H3K9me2 or H3K81ac were independent predictors of poor survival. If a low cellular level of both H3K4me2 and H3K9me2 was observed, the survival outcome was even worse.10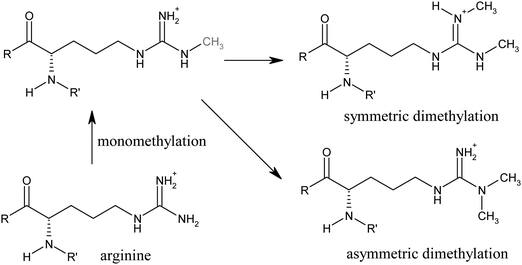 | ||
| Fig. 1 Arginine methylation. | ||
For a long time, histone methylation was conceived to be a permanent mark11 due to the half-life of methylated lysines that is comparable to the half-life of histones.12 The first discovery of a demethylating activity by Paik and coworkers13 in 1973 describes that an extract obtained from rat kidney was able to convert mono- and dimethylated lysine residues to their unmodified status. Shi et al. were the first to report about a histone demethylase in 2004, namely LSD1, that is able to demethylate H3K4 me1/me2.14 In contrast, methyl groups from trimethylated lysines could not be cleaved off by LSD1. Instead, JmjC-domain-containing enzymes were found to demethylate trimethylated lysines.15–17 Monomethylated arginine residues can be deiminated to citrulline by PAD4, a peptidylarginine deiminase.18 Dimethylated arginines in contrast, have been reported to be directly converted to arginine by the demethylase JMJD6.19 The latter has been discussed controversially lately, as Webby and coworkers have shown a lysyl-hydroxylation activity of JMJD6 but they could not observe an argininedemethylation.20 The therapeutic potential of drugs that influence posttranslational modifications is vast as there are many diseases, among them especially cancer, that have been linked to aberrant gene expression patterns or deregulated histone modifying enzymes.21
2.2 Non-histone protein methylation
Besides histoneproteins, many other targets for histonemethyltransferases have been identified so far. To name just a few, argininemethylation of FOXOtranscription factors by PRMT1 (proteinargininemethyltransferase 1) inhibits their phosphorylation by Akt which in turn leads to an increase in oxidative-stress-induced apoptosis mediated by the PI3K-Akt signalling pathway.22 Moreover, PIAS1, a STAT1 inhibitor in the IFN-γsignalling pathway, is methylated by PRMT1 which has an impact on STAT1 target gene expression.23 Furthermore, p53 is subject to methylation. Once methylated by PRMT5 on arginines R333, R335 and R337, which are part of the oligomerization domain, target gene specifity is affected.24 PRMT5 depletion leads to apoptosis because of an altered promoter binding specifity of p53.24 Upon monomethylation of p53 at lysine 382 by Set8 (KMT5A) transcriptional activation by p53 is suppressed. Here, knockdown of Set8 resulted in increased apoptosis.25 When, in contrast, p53 is methylated on lysine 372 by Set9 (KMT7) within the C-terminus, p53 is stabilized and p53-dependent transcription is augmented.26Interestingly, the HIV transactivator protein Tat is subject both to lysinemethylation by SETDB1 (KMT1E), SETDB2 (KMT1F)27 and Set7/9 (KMT7)28 and argininemethylation by PRMT6.29 The latter leads to negative regulation of the transactivation activity.29Lysinemethylation of TatK50 and TatK51 by SETDB1 resp. SETDB2 seems to prevent lysineacetylation of the same residues which in turn prevents viral transactivation.27 On the other hand, data of Pagans et al. indicate that monomethylation of TatK51 by Set7/9 is coactivating Tat transactivation.28 Within the histoneproteins, lysinemethyltransferases are usually highly site-specific as compared to e.g. HDACs.30 But as to their general substrate specificity, it has been shown e.g. for G9a that it methylates the lysines of many protein substrates and not only histones.31
3. Histone methyltransferases and their inhibitors
Histone methyltransferases can be subdivided into three classes: SET-domain containing lysinemethyltransferases, non-SET domain lysinemethyltransferases and arginine methyltransferases (PRMTs). All of them use S-adenosylmethionine (SAM) as a co-substrate for the methylation and S-adenosylhomocysteine (SAH) is formed as a byproduct.32 Unlike histone acetylation, histone methylation does not alter the charge of the lysine residue in question. It affects the basicity, hydrophobicity and the size of the amino acid side chain group which affects the affinity of proteins that recognize such side chains. Although these changes are subtle, there are proteins that are able to bind selectively to certain methylated residues. These proteins have so-called chromodomains that recognize the methylated lysines. The structural data available led to an understanding of the motifs involved in methyl lysine binding.33 Chromodomains recognize mono-, di- or trimethylated lysine residues. Tudor domains bind trimethylated lysines as well but there is also evidence for the recognition of mono- and dimethylated H4K20. PHD (plant homeo domain) fingers preferentially bind di- or trimethylated lysines. There are MBT (malignant brain tumor) domains that specifically bind mono-and dimethylated lysines. The only known proteins that recognize methylated arginines are the so-called WD40 repeats. They consist of about 40 amino acids and often have a tryptophan - aspartic acid dipeptide (W–D) at the C-terminus. WD40 repeats can bind dimethylated arginine, namely H3R2(me)2 but also dimethylated lysines.33–36So far, more than 20 lysinemethyltransferases37 and eleven arginine methyltransferases (PRMT1-11) have been identified, but not for all of the members enzymatic activity could be demonstrated.38–40Lysine methyltransferases have been designated historically with a variety of names. Newly, a common nomenclature for chromatin modifying enzymes has been proposed instead of these. The human lysine methyltransferases have been renamed KMTs and are subdivided into 8 classes.41 Many of the methyltransferases have been linked to cancer and therefore the development of inhibitors is desirable.42
3.1 Lysine methyltransferases
Lysine methyltransferases consist of two classes - lysinemethyltransferases with or without a SET-domain.32 As already mentioned, the biological consequences of lysinemethylation depend on the extent of methylation and the target site. Additionally, the methylation may lead to further biochemical changes in a cellular context and this may have varying consequences for different forms of diseases as described below (see Table 1).34Opposing effects have been discovered concerning the outcome of lysinemethylation: methylation of the tumor suppressor protein p53 on K370 by Smyd2 (KMT3C)43 or on K382 by SET8 (KMT5A)25 leads to inhibition of transcriptional activity whereas methylation on K372 by Set9 (KMT7) results in transcriptional activation.26 The lysinemethyltransferase G9a (KMT1C) has recently been shown to dimethylate p53 on K373. This was correlated with inactivity of p53.44 Its impact on apoptotic processes as well as its overexpression in various cancer types suggests that G9a is a putative oncogene.44 Interestingly, another study revealed a correlation between cocaine addiction and cellular levels of the lysinemethyltransferase G9a with the conclusion that reduced levels of G9a expression and subsequent hypomethylation on H3K9 correlate with cocaine addiction. Repeated administration of cocaine in mice showed a downregulation of G9a. G9a overexpression in mice by direct injections into the Nucleus accumbens with Herpes simplex virus vectors expressing G9a reduced the preference for cocaine. These insights may help to develop more effective treatments for drug addiction and an improved understanding of the biochemical basis of drug addiction.45
As causes for dysregulated histone methylation, the enzyme in question might be mutated or overexpressed, e.g. in cancer cells. The lysinemethyltransferase EZH2 (KMT6) was found to be increased in breast and prostate cancer46 with a poor prognosis for elevated levels in prostate cancer.47 Mutations of the methyltransferase MLL1 seem to be responsible for various forms of acute leukaemia by blocking hematopoietic differentiation.48 Hypermethylation of H3K79 by the associated complex MLL-AF10 or MLL-hDOT1L respectively is reported to result in leukaemogenesis.49 Additionally, SMYD3 was found to be upregulated in colorectal, hepatocellular50 and breast cancer. For the latter, overexpression led to enhanced cancer cell growth.51 Furthermore, SET7/9 (KMT7) has recently been associated with hyperglycaemia-induced epigenetic changes. Transient high glucose levels seem to result in SET7/9 methylating histones in the promoter region of nuclear factor κB (NF-κB) causing increased p65 gene expression.52
| Epigenetic protein | Links to disease | Major target sitesa |
|---|---|---|
| a If no reference is cited, source is Ref. 41. | ||
| SUV39H1 (KMT1A) | Elevated mRNA level in colon cancer tissue samples94 | H3K9 |
| G9a (KMT1C) | Suppressor gene silencing95 | H3K9 |
| Eu-HMTase1 (KMT1D) | Overexpression in gland tumors96 | H3K9 |
| MLL1 (KMT2A) | Rearrangement/amplification blocks hematopoietic differntiation48 | H3K4 |
| MLL4 (KMT2D) | Involved in hepatitis B virus dependent liver carcinogenesis97 | H3K4 |
| SMYD2 (KMT3C) | Suppression of p53 transcriptional activity43 | H3K56, p53K370 |
| SMYD3 | Overexpression and enhanced tumor cell growth in breast cancer51 | H3K450 |
| Overexpression in colon cancer and hepatocellular carcinoma50 | ||
| DOT1L (KMT4) | Enzymatic activity crucial for leukaemogenesis49 | H3K79 |
| EZH2 (KMT6) | Amplification and overexpression in multiple cancer types98 | H3K27 |
| SET7/9 (KMT7) | Hyperglycemia induces SET7/9 causing increased p65 gene expression.52 | H3K4, p53K37226 |
3.2 Inhibitors of lysine methyltransferases
Generally, methyltransferases can be inhibited by co-substrate analogues. There are three known co-substrate analogues that inhibit a variety of methyltransferases: sinefungin, an antibiotic compound that is structurally similar to SAM, the demethylated co-substrate SAH as a feedback inhibitor, and methylthioadenosine. This kind of inhibitors is rather unselective and as such not applicable as therapeutic agents.53There are only a few drug-like inhibitors of lysinemethyltransferases and most of them were discovered by random screening approaches. The first inhibitor was chaetocin (1, see Fig. 2) which is a fungal mycotoxin. It inhibits both the Drosophila melanogasterenzyme Su(var)3–9 with an IC50 of 0.8 μM and G9a with an IC50 of 2.5 μM.54 Recently, chaetocin was also shown to be highly active against myeloma cells by increasing the sensitivity to oxidative stress induced cytotoxicity with hardly any effect on normal human B cells.55
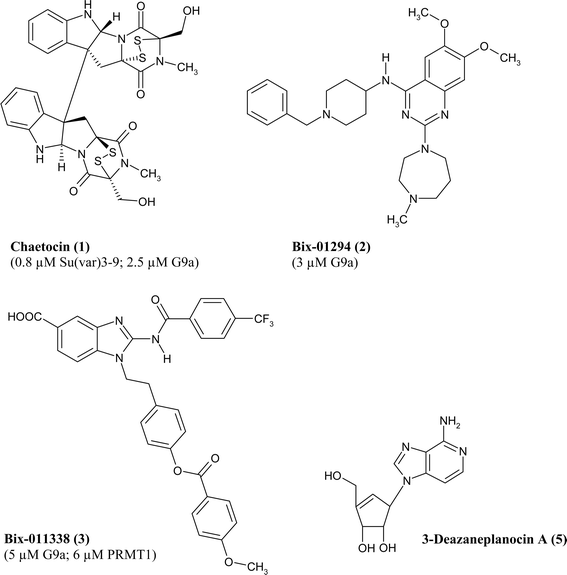 | ||
| Fig. 2 Lysine methyltransferase inhibitors (IC50 values and corresponding enzymes in parentheses). | ||
The compound Bix-01294 (2, see Fig. 2) was identified in a combined virtual and high-throughput screen approach and shown to be an inhibitor of G9a (KMT1C) with an IC50 of 3 μM. It is selective against SUV39H1 and PRMT1. Cultured cells treated with Bix-01294 showed a reduction of histone H3K9 dimethylation and a decrease in cell number. In the same screening procedure, the compound Bix-01338 (3, see Fig. 2) was discovered as a rather unselective inhibitor of methyltransferases. It did not show any selectivity between lysine or argininemethyltransferases with an IC50 of 5 μM for G9a and an IC50 of 6 μM for PRMT1.56
Recently, UNC0224 (4, see Fig. 3) has been presented as a new inhibitor for the lysinemethyltransferase G9a with an IC50 of 15 nM. This 2,4-diamino-7-aminoalkoxyquinazoline has been found by modifying the structure of Bix-01294. It showed a 1000-fold selectivity for G9a compared to SET7/9 (KMT7) and Set8. Additionally, the first crystal structure of G9a with an inhibitor in a complex was introduced with Bix-01294 (see Fig. 4).57Bix-01294 forms two hydrogen bonds to Asp1131 and Asp1140 at the entrance of the substrate binding pocket, whereas the two methoxy groups show van der Waals interactions with the more buried part of the pocket. Interestingly, the inhibitor is not involved in direct interaction with the residues of the catalytic site or the cofactorSAM. Therefore, the dimethylaminopropoxy side chain in UNC0224 was attached in order to obtain binding to the lysine tunnel and hence increased inhibitory activity. The structural information is particularly important because of the known link of G9a to human cancers.44,58
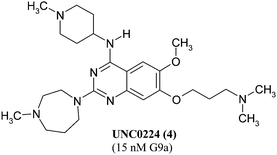 | ||
| Fig. 3 Lysine methyltransferase inhibitor obtained by structural variation of BIX-01294 (IC50 value and corresponding enzyme in parentheses). | ||
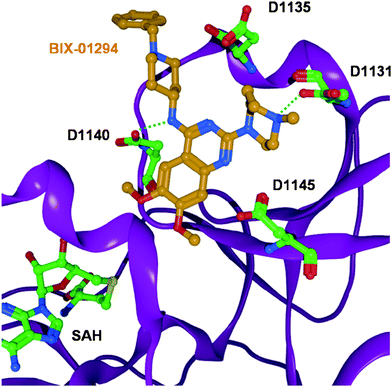 | ||
| Fig. 4 Crystal structure of the G9a methyltransferase in complex with the inhibitorBix-01294 (coloured orange). The inhibitor is involved in two hydrogen bonds to D1131 and D1140. (The cofactor analogue SAH is coloured green and the protein backbone is shown as purple ribbon. Only interacting residues of G9a are displayed). | ||
A new combined epigenetic therapy has been proposed using the EZH2 (KMT6) inhibitor3-deazaneplanocin A (DZNep, 5, see Fig. 2) and the histone deacetylase inhibitorpanobinostat.59 EZH2 is a part of the polycomb repressive complex 2 with lysinemethyltransferase activity and is involved in proliferation and aggressive cell growth. DZNep is a cyclopentenyl analogue of 3-deazaadenosine and was originally synthesized as an inhibitor of S-adenosyl-L-homocysteine hydrolase.60 DZNep treatment of cultured human acute myeloid leukaemia cells induced an increased expression of p16, p21, p27 and FBXO32 as well as apoptosis. These effects were enhanced by cotreatment with the pan-HDAC inhibitorpanobinostat. Also the survival of NOD/SCID mice with leukaemia caused by the AML HL-60 cells was improved upon cotreatment.59
3.3 Arginine methyltransferases
Protein arginine methylation is mediated by PRMTs that can be subdivided into two classes: type I methyltransferases catalyse the formation of asymmetrically substituted arginine residues and type II methyltransferases mediate the formation of symmetrically methylated arginine residues. PRMTs 1–4, 6 and 8 are type I enzymes and PRMTs 5, 7 and 9 are PRMTs of type II (see Table 2). PRMTs 2 and 9 were shown to be catalytically inactive and for PRMTs 10 and 11 a proof of methyltransferase activity is lacking so far as well.61 PRMTs usually methylate glycine-alanine-arginine patches (so-called GAR motifs). As an exception, CARM1 shows affinity to proline-glycine-methionine-arginine sequences (so-called PGM motifs).39 PRMT5 has also been shown to methylate PGM motifs.62PRMT1 is an essential component of the Mixed Lineage Leukaemia (MLL) oncogenic transcriptional complex. It has transcriptional activation properties when aberrantly expressed and hence is critical for the development of leukemia.63 As quoted above, PRMT1 has also been shown to methylate FOXOtranscription factors that play a role in the homoeostasis of cellapoptosis. By methylationvia PRMT1, the FOXOproteins cannot be further phosphorylated by Akt. This leads to an increase in oxidative-stress-induced apoptosis which is based upon the PI3K-Akt signalling pathway.22 Here, inhibitors of PRMT1 could be beneficial for the therapy of neurodegenerative diseases as one major issue of it is cell damage due to oxidative stress. Methylation of a conserved arginine residue in the Igα subunit of the B cellreceptor by PRMT1 shows a role in immunology as it promotes B cell differentiation.64
Arginine methyltransferases are known to be co-activators of nuclear receptors and therefore an interesting target for the therapy of hormone dependent cancer.65,66E.g., PRMT2 and 5 have been identified as co-activators of the androgen receptor. For PRMT1, 2 and 4 (CARM1) there is evidence of being estrogen receptor associated co-activators. Both PRMT1 and CARM1 seem to stimulate estrogen receptor mediated transcription by interacting with members of the p160 family.67 Furthermore, the histone acetyltransferaseCREB bindingprotein (CBP) is an important regulator for the transcriptional activation by nuclear receptors and other transcription factors. There is evidence that CBP is methylated by CARM1 which enhances its co-activating activity.68
PRMT5 appears to be a regulator of p53 transcription. In the context of DNA damage control reduced levels of PRMT5 lead to apoptosis with an altered p53 promoter binding specifity indicating the regulatory function of PRMT5.24 Additionally, PRMT5 has been connected to proliferation and regulation of cell growth by negatively controlling the expression of tumor suppressor genes ST7 and NM23.69
PRMT7 is also a potential target of cancer therapy because downregulation of PRMT7 seems to sensitize cancer cells to camptothecin treatment.70 Thus, inhibitors of PRMT7 could be used to potentiate the effects of this and maybe other cytotoxic drugs.
Arginine methylation activity can be regulated viaPRMT-binding proteins such as BTG1 and TIS2/BTG2,71 negative feedback by SAH or posttranslational modifications.39E.g., it has been shown that phosphorylation of CARM1 negatively regulates its arginine methylating activity. Different serine residues have been identified as potential phosphorylation sites whereas the responsible kinase is still unknown.72,73
Arginine methylation is also involved in RNA maturation62 and DNA repair.74 The checkpoint protein MRE11 plays an important role in DNA damage control and is dependent on argininemethylation75 implying a possible safety problem for using argininemethyltransferaseinhibitors as drugs. Nevertheless, there is a great therapeutic potential for inhibitors of argininemethyltransferases for a variety of diseases.
| Epigenetic protein | Links to disease | Major target sitesa |
|---|---|---|
| a If no reference is cited, source is Ref. 61. | ||
| PRMT1 | Essential component of MLL oncogenic transcriptional complex63 | H4R3, RG domains of Npl399 |
| Coactivator of hormonereceptor function76 | ||
| PRMT2 | Androgen receptor coactivator100 | No catalytic activity |
| PRMT4 (CARM1) | Overexpression in breast cancer101 | H3R2, H3R17, H3R26, CBP/p300R2142103 |
| Overexpression correlates with androgen independence in prostate tumor cells102 | ||
| Contribution to CBP coactivation due to methylation68 | ||
| PRMT5 | Overexpression in lymphoid cancer cells104 | H3R8, H4R3, RG domain of MBD2105,106 |
| PRMT6 | Overexpression diminished viral Tat activity29 | H2AR3, H3R2, H4R3107 |
| PRMT7 | Sensitization of tumor cells to camptothecin treatment when downregulated70 | Not known |
3.4 Inhibitors of arginine methyltransferases
As seen for lysine methyltransferases, co-substrate analogues like sinefungin can be used as inhibitors of argininemethyltransferases with the same restrictions as mentioned above.The first more specific inhibitors of argininemethyltransferases were discovered in a random screening approach. The screening was performed with the yeast enzyme Hmt1p and with Npl3 as a nonhistone substrate. Nine of the 9000 compounds screened showed a good activity in the low micromolar range and were then tested against human PRMT1. The new inhibitors were called AMIs for argininemethyltransferaseinhibitors. AMI-1 was the most potent inhibitor with an IC50 of 9 μM against PRMT1 (6, see Fig. 5). It further showed selectivity towards several lysinemethyltransferases and in vivo effects in a cell culture model. The methylation level of Npl3 in HeLa cells that were transiently transfected with GFP-Npl3 was decreased upon AMI-1 treatment and an inhibitory effect on the activation on nuclear androgen and estrogen receptors could be shown.76 As a caveat, AMI-2 contains partial structures well known for promiscuous effects like aggregation and some of the AMIin vitro results have not yet been verified in cell culture.77AMI-1 shows resemblance to the antiparasitic drugsuramin and similarly inhibits sirtuins, yet with decreased activity (IC50 on Sirt1: 300 nM for suramin, 32 μM for AMI-1).78
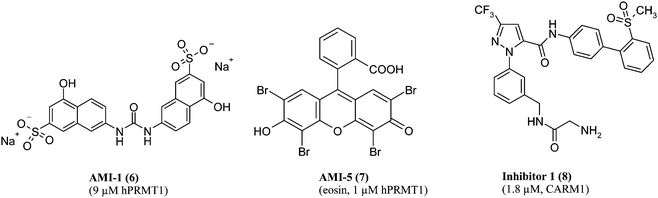 | ||
| Fig. 5 Arginine methyltransferase inhibitors obtained from random screening (IC50 value and corresponding enzyme in parentheses). | ||
Among the AMI compounds, AMI-5, (eosin, 7, see Fig. 5) has been used as lead structure to synthesize further inhibitors of methyltransferases. One example is compound 6e (15, see Fig. 6).79 It was subsequently modified to give simplified scaffolds that are structurally similar to curcumin including bromo- or dibromophenol substructures. Compound 8e (16, see Fig. 6) was selective for CARM1 over PRMT1 at a concentration of 100 μM.80 The newly obtained PRMTinhibitors were also tested against HAT and sirtuins due to the fact that curcumin is a known HATinhibitor81 and the compound GW5074 with a dibromophenol substructure exhibits sirtuin inhibitory properties.82 Both HAT and sirtuin inhibitory activity was confirmed in vitro for several compounds. Cellular hypomethylation was demonstrated for some inhibitors but data on cellular hypoacetylation is not available.
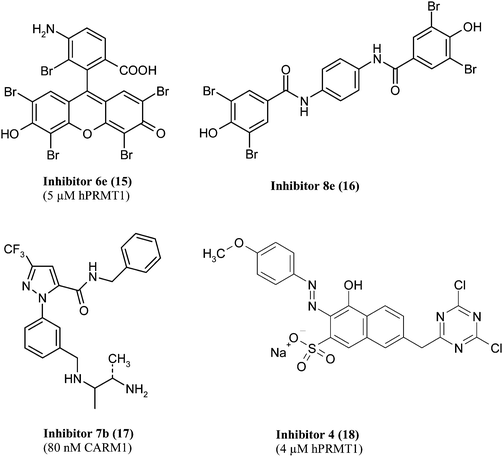 | ||
| Fig. 6 Arginine methyltransferase inhibitors obtained by synthetic variation of screening hits (IC50 value and corresponding enzyme in parentheses). | ||
The aminonaphthol sulfonate core of AMI-1 was used to develop further potent and selective inhibitors for PRMT1 by Bonham and co-workers.83 They combined the structural core with the azo moiety of AMI-976 and the dichlorotriazinegroup of AMI-676 on the other hand to give Inhibitor 4 (18, see Fig. 6, IC50 = 4 μM). As PRMT1 activity is involved in T helper cell function84–86 the inhibitory effect of Inhibitor 4 towards T helper cellcytokine production was characterized. Inhibitor 4 enhanced T-helper cell proliferation. Additionally, decreased IFN-γ levels of type 1 T-helper cells and IL-4 levels of type 2 T-helper cells were observed upon inhibitor treatment. Thus, this new compound shows potential in the treatment of autoimmune diseases.
A further compound called inhibitor1 (8, see Fig. 5) which is selective towards CARM1 and has been obtained from high-throughput screening was further modified to give compound 7b (17, see Fig. 6). 7b showed an IC50 of 80 nM and selectivity towards other methyltransferases.87
The first target based virtual screening for argininemethyltransferases was presented by our group in 2007 using a homology model of hPRMT1.88 The screening procedure was created in a way that potential ligands for the substrate binding pocket were selected but not those that would target the co-substrate binding site. About 40 compounds of the NCI diversity set were selected for in vitro testing by manually inspecting the screening hits. Seven inhibitors with an IC50 below 60 μM were found in the subsequent in vitro assay. The inhibitorsallantodapsone (9, see Fig. 7) and stilbamidine (10, see Fig. 7) induced hypomethylation at H4R3 in HepG2 cells. Functional activity was confirmed in a reporter gene assay with MCF7a cells. Both inhibitors showed a reduction of estrogen receptoractivation by estradiol in a dose-dependent manner.88 Competition studies using histone substrate and cofactor SAM showed that the identified inhibitors are substrate competitive inhibitors, which is in agreement with the obtained docking results. In Fig. 8 the predicted binding mode of allantodapsone and stilbamidine is presented. Both are making hydrogen bonds to the acidic residue (E152, E161) of the catalytic site.
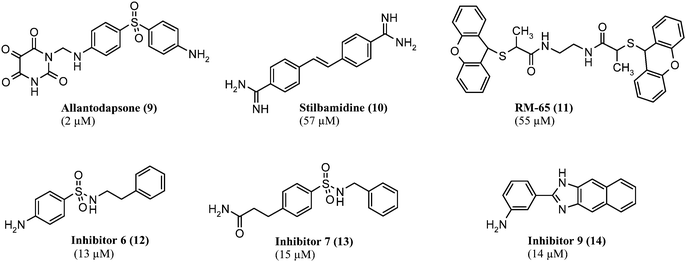 | ||
| Fig. 7 Arginine methyltransferase inhibitors from virtual screening (IC50 value against hPRMT1 in parentheses). | ||
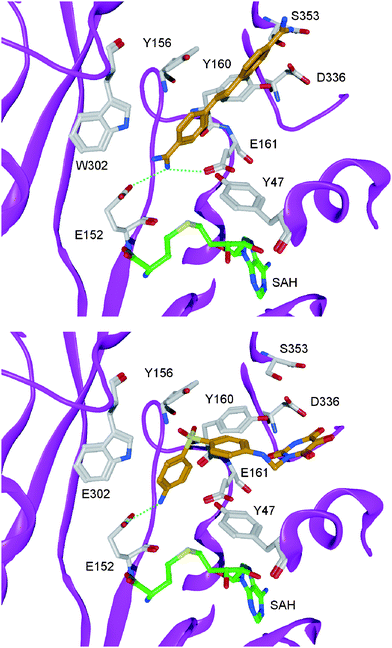 | ||
| Fig. 8 Docking solutions for the PRMT1 inhibitorsstilbamidine (top, coloured orange) and allantodapsone (bottom, coloured orange). Hydrogen bonds are shown as dashed lines, the protein backbone is shown as pink ribbon and the cofactor analogue SAH is coloured green. | ||
As a second approach, a fragment-based virtual screening for molecules with inhibitory properties and a molecular weight below 200 g mol−1 was performed leading to the discovery of an α–methylthioglycolic amide as an inhibitor for PRMT1. As this compound exhibited neither good chemical stability nor a drug-like structure, a similarity search was conducted. As a result, the compound RM-65 (11, see Fig. 7) could be identified as inhibitor of PRMT1 in vitro with cellular hypomethylating activity in HepG2 cells at the PRMT1 target H4R3. It does not inhibit the lysinemethyltransferase SET7/9. Although the original search was performed with fragments binding to the substrate binding pocket, RM-65 rather seems to be a bisubstrate mimic as proposed by a docking study on a hPRMT1 homology model.89
In the further course of research, greater databases were screened for new argininemethyltransferaseinhibitors with a modified screening procedure including a pharmacophore prefilter. This approach conduced to the discovery of inhibitors6, 7 and 9 (12, 13, 14 see Fig. 7) from the Chembridge database with inhibitory effects down to 13 μM.90
4. Conclusion
For methyltransferases many links to the pathogenesis of various diseases have been demonstrated. Thus, the further development of inhibitors is extremely important for the future therapy of the various diseases, especially for cancer. The major issue that needs to be addressed is the selectivity among the different histonemethyltransferases in order to better understand the function of the individual subtypes which should eventually lead to optimized therapeutics. Recently obtained structural information should be important information towards obtaining subtype selectivity.91Equally important is a better understanding of the relevance of histonevs.non-histonemethylation for the resulting phenotype.Even though the field of epigenetic drug discovery is still at its beginning, a dynamic development can be anticipated and clearly further epigenetic drugs based on histonemethyltransferases will be developed in the near future. Of course, also the histone demethylases will be similarly in the focus. Special attention will also be given to the methyl binding proteins, the so-called readers of the histone code. As screening assays have become available for the testing of this recognition,92,93inhibitors of this interaction are to be expected soon which may open up a completely new class of epigenetic therapeutics.
5. Acknowledgements
We would like to thank the Deutsche Krebshilfe for financial support of our work on PRMT inhibitors.6. References
- S. Schafer and M. Jung, Chromatin modifications as targets for new anticancer drugs, Arch. Pharm., 2005, 338, 347–357 CrossRef.
- C. H. Henkels and S. Khorasanizadeh, Implications of a histone code mimic in epigenetic signaling, Mol. Cell, 2007, 27, 521–522 CrossRef CAS.
- M. A. Holbert and R. Marmorstein, Structure and activity of enzymes that remove histone modifications, Curr. Opin. Struct. Biol., 2005, 15, 673–680 CrossRef CAS.
- T. Suzuki and N. Miyata, Epigenetic control using natural products and synthetic molecules, Curr. Med. Chem., 2006, 13, 935–958 CrossRef CAS.
- Y. G. Zheng, J. Wu, Z. Chen and M. Goodman, Chemical regulation of epigenetic modifications: opportunities for new cancer therapy, Med. Res. Rev., 2008, 28, 645–687 CrossRef CAS.
- A. Mai and L. Altucci, Epi-drugs to fight cancer: from chemistry to cancer treatment, the road ahead, Int. J. Biochem. Cell Biol., 2009, 41, 199–213 CrossRef CAS.
- A. Spannhoff, A. T. Hauser, R. Heinke, W. Sippl and M. Jung, The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors, ChemMedChem, 2009, 4, 1568–1582 CrossRef CAS.
- C. Martin and Y. Zhang, The diverse functions of histone lysine methylation, Nat. Rev. Mol. Cell Biol., 2005, 6, 838–849 CrossRef CAS.
- M. F. Fraga, E. Ballestar, A. Villar-Garea, M. Boix-Chornet and J. Espada, et al., Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer, Nat. Genet., 2005, 37, 391–400 CrossRef CAS.
- A. Manuyakorn, R. Paulus, J. Farrell, N. A. Dawson and S. Tze, et al., Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704, J. Clin. Oncol., 2010, 28, 1358–1365 CrossRef CAS.
- J. C. Rice and C. D. Allis, Histone methylation versus histone acetylation: new insights into epigenetic regulation, Curr. Opin. Cell Biol., 2001, 13, 263–273 CrossRef CAS.
- J. A. Duerre and C. T. Lee, In vivo methylation and turnover of rat brain histones, J. Neurochem., 1974, 23, 541–547 CrossRef CAS.
- W. K. Paik and S. Kim, Enzymatic demethylation of calf thymus histones, Biochem. Biophys. Res. Commun., 1973, 51, 781–788 CrossRef CAS.
- Y. Shi, F. Lan, C. Matson, P. Mulligan and J. R. Whetstine, et al., Histone demethylation mediated by the nuclear amine oxidase homolog LSD1, Cell, 2004, 119, 941–953 CrossRef CAS.
- K. Agger, P. A. Cloos, J. Christensen, D. Pasini and S. Rose, et al., UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development, Nature, 2007, 449, 731–734 CrossRef CAS.
- Y. Xiang, Z. Zhu, G. Han, H. Lin and L. Xu, et al., JMJD3 is a histone H3K27 demethylase, Cell Res., 2007, 17, 850–857 CrossRef CAS.
- D. J. Seward, G. Cubberley, S. Kim, M. Schonewald and L. Zhang, et al., Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins, Nat. Struct. Mol. Biol., 2007, 14, 240–242 CrossRef CAS.
- P. R. Thompson and W. Fast, Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock?, ACS Chem. Biol., 2006, 1, 433–441 CrossRef CAS.
- B. Chang, Y. Chen, Y. Zhao and R. K. Bruick, JMJD6 is a histone arginine demethylase, Science, 2007, 318, 444–447 CrossRef CAS.
- C. J. Webby, A. Wolf, N. Gromak, M. Dreger and H. Kramer, et al., Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing, Science, 2009, 325, 90–93 CrossRef CAS.
- M. Esteller, CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future, Oncogene, 2002, 21, 5427–5440 CrossRef CAS.
- K. Yamagata, H. Daitoku, Y. Takahashi, K. Namiki and K. Hisatake, et al., Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt, Mol. Cell, 2008, 32, 221–231 CrossRef CAS.
- S. Weber, F. Maass, M. Schuemann, E. Krause and G. Suske, et al., PRMT1-mediated arginine methylation of PIAS1 regulates STAT1 signaling, Genes Dev., 2009, 23, 118–132 CrossRef CAS.
- M. Jansson, S. T. Durant, E. C. Cho, S. Sheahan and M. Edelmann, et al., Arginine methylation regulates the p53 response, Nat. Cell Biol., 2008, 10, 1431–1439 CrossRef CAS.
- X. Shi, I. Kachirskaia, H. Yamaguchi, L. E. West and H. Wen, et al., Modulation of p53 function by SET8-mediated methylation at lysine 382, Mol. Cell, 2007, 27, 636–646 CrossRef CAS.
- S. Chuikov, J. K. Kurash, J. R. Wilson, B. Xiao and N. Justin, et al., Regulation of p53 activity through lysine methylation, Nature, 2004, 432, 353–360 CrossRef CAS.
- R. Van Duyne, R. Easley, W. Wu, R. Berro and C. Pedati, et al., Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR, Retrovirology, 2008, 5, 40 CrossRef.
- S. Pagans; S. E. Kauder; K. Kaehlcke; N. Sakane; S. Schroederet al., The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription. Cell Host Microbe, 7, pp. 234–244 Search PubMed.
- M. C. Boulanger, C. Liang, R. S. Russell, R. Lin and M. T. Bedford, et al., Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression, J. Virol., 2005, 79, 124–131 CrossRef CAS.
- M. Tachibana, K. Sugimoto, T. Fukushima and Y. Shinkai, Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3, J. Biol. Chem., 2001, 276, 25309–25317 CrossRef CAS.
- P. Rathert, A. Dhayalan, M. Murakami, X. Zhang and R. Tamas, et al., Protein lysine methyltransferase G9a acts on non-histone targets, Nat. Chem. Biol., 2008, 4, 344–346 CrossRef CAS.
- B. C. Smith and J. M. Denu, Chemical mechanisms of histone lysine and arginine modifications, Biochim. Biophys. Acta, 2009, 1789, 45–57 CAS.
- P. R. Nielsen, D. Nietlispach, H. R. Mott, J. Callaghan and A. Bannister, et al., Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9, Nature, 2002, 416, 103–107 CrossRef CAS.
- A. J. Ruthenburg, H. Li, D. J. Patel and C. D. Allis, Multivalent engagement of chromatin modifications by linked binding modules, Nat. Rev. Mol. Cell Biol., 2007, 8, 983–994 CrossRef CAS.
- S. A. Jacobs and S. Khorasanizadeh, Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail, Science, 2002, 295, 2080–2083 CrossRef CAS.
- E. J. Neer, C. J. Schmidt, R. Nambudripad and T. F. Smith, The ancient regulatory-protein family of WD-repeat proteins, Nature, 1994, 371, 297–300 CrossRef CAS.
- C. Qian and M. M. Zhou, SET domain protein lysine methyltransferases: Structure, specificity and catalysis, Cell. Mol. Life Sci., 2006, 63, 2755–2763 CrossRef CAS.
- F. M. Boisvert, C. A. Chenard and S. Richard, Protein interfaces in signaling regulated by arginine methylation, Science's STKE, 2005, 2005, re2 Search PubMed.
- M. T. Bedford, Arginine methylation at a glance, J. Cell Sci., 2007, 120, 4243–4246 CrossRef CAS.
- S. Pal and S. Sif, Interplay between chromatin remodelers and protein arginine methyltransferases, J. Cell. Physiol., 2007, 213, 306–315 CrossRef CAS.
- C. D. Allis, S. L. Berger, J. Cote, S. Dent and T. Jenuwien, et al., New nomenclature for chromatin-modifying enzymes, Cell, 2007, 131, 633–636 CrossRef CAS.
- R. Schneider, A. J. Bannister and T. Kouzarides, Unsafe SETs: histone lysine methyltransferases and cancer, Trends Biochem. Sci., 2002, 27, 396–402 CrossRef CAS.
- J. Huang, R. Sengupta, A. B. Espejo, M. G. Lee and J. A. Dorsey, et al., p53 is regulated by the lysine demethylase LSD1, Nature, 2007, 449, 105–108 CrossRef CAS.
- J. Huang; J. Dorsey; S. Chuikov; X. Zhang; T. Jenuweinet al., G9A and GLP methylate lysine 373 in the tumor suppressor p53. J Biol Chem Search PubMed.
- I. Maze; H. E. Covington, 3rd; D. M. Dietz; Q. LaPlant; W. Renthalet al., Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science, 327, pp. 213–216 Search PubMed.
- C. S. Cooper, C. Campbell and S. Jhavar, Mechanisms of Disease: biomarkers and molecular targets from microarray gene expression studies in prostate cancer, Nat. Clin. Pract. Urol., 2007, 4, 677–687 CAS.
- R. J. Bryant, N. A. Cross, C. L. Eaton, F. C. Hamdy and V. T. Cunliffe, EZH2 promotes proliferation and invasiveness of prostate cancer cells, Prostate, 2007, 67, 547–556 CrossRef CAS.
- J. L. Hess, Mechanisms of transformation by MLL, Crit. Rev. Eukaryotic Gene Expression, 2004, 14, 235–254 Search PubMed.
- Y. Okada, Q. Feng, Y. Lin, Q. Jiang and Y. Li, et al., hDOT1L links histone methylation to leukemogenesis, Cell, 2005, 121, 167–178 CrossRef CAS.
- R. Hamamoto, Y. Furukawa, M. Morita, Y. Iimura and F. P. Silva, et al., SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells, Nat. Cell Biol., 2004, 6, 731–740 CrossRef CAS.
- R. Hamamoto, F. P. Silva, M. Tsuge, T. Nishidate and T. Katagiri, et al., Enhanced SMYD3 expression is essential for the growth of breast cancer cells, Cancer Sci., 2006, 97, 113–118 CrossRef CAS.
- A. El-Osta, D. Brasacchio, D. Yao, A. Pocai and P. L. Jones, et al., Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia, J. Exp. Med., 2008, 205, 2409–2417 CrossRef CAS.
- M. Vedel, F. Lawrence, M. Robert-Gero and E. Lederer, The antifungal antibiotic sinefungin as a very active inhibitor of methyltransferases and of the transformation of chick embryo fibroblasts by Rous sarcoma virus, Biochem. Biophys. Res. Commun., 1978, 85, 371–376 CrossRef CAS.
- D. Greiner, T. Bonaldi, R. Eskeland, E. Roemer and A. Imhof, Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9, Nat. Chem. Biol., 2005, 1, 143–145 CrossRef CAS.
- C. R. Isham, J. D. Tibodeau, W. Jin, R. Xu and M. M. Timm, et al., Chaetocin: a promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress, Blood, 2007, 109, 2579–2588 CrossRef CAS.
- S. Kubicek, R. J. O'Sullivan, E. M. August, E. R. Hickey and Q. Zhang, et al., Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase, Mol. Cell, 2007, 25, 473–481 CrossRef CAS.
- F. Liu, X. Chen, A. Allali-Hassani, A. M. Quinn and G. A. Wasney, et al., Discovery of a 2,4-Diamino-7-aminoalkoxyquinazoline as a Potent and Selective Inhibitor of Histone Lysine Methyltransferase G9a, J. Med. Chem., 2009 Search PubMed.
- Y. Kondo, L. Shen, S. Ahmed, Y. Boumber and Y. Sekido, et al., Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells, PLoS One, 2008, 3, e2037 CrossRef.
- W. Fiskus, Y. Wang, A. Sreekumar, K. M. Buckley and H. Shi, et al., Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells, Blood, 2009, 114, 2733–2743 CAS.
- R. I. Glazer, K. D. Hartman, M. C. Knode, M. M. Richard and P. K. Chiang, et al., 3-Deazaneplanocin: a new and potent inhibitor of S-adenosylhomocysteine hydrolase and its effects on human promyelocytic leukemia cell line HL-60, Biochem. Biophys. Res. Commun., 1986, 135, 688–694 CrossRef CAS.
- C. D. Krause, Z. H. Yang, Y. S. Kim, J. H. Lee and J. R. Cook, et al., Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential, Pharmacol. Ther., 2007, 113, 50–87 CrossRef CAS.
- D. Cheng, J. Cote, S. Shaaban and M. T. Bedford, The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing, Mol. Cell, 2007, 25, 71–83 CrossRef CAS.
- N. Cheung, L. C. Chan, A. Thompson, M. L. Cleary and C. W. So, Protein arginine-methyltransferase-dependent oncogenesis, Nat. Cell Biol., 2007, 9, 1208–1215 CrossRef CAS.
- S. Infantino; B. Benz; T. Waldmann; M. Jung; R. Schneideret al., Arginine methylation of the B cell antigen receptor promotes differentiation. J Exp Med, 207, pp. 711–719 Search PubMed.
- M. T. Bedford and S. Richard, Arginine methylation an emerging regulator of protein function, Mol. Cell, 2005, 18, 263–272 CrossRef CAS.
- C. Qi, J. Chang, Y. Zhu, A. V. Yeldandi and S. M. Rao, et al., Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha, J. Biol. Chem., 2002, 277, 28624–28630 CrossRef CAS.
- D. Chen, S. M. Huang and M. R. Stallcup, Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300, J. Biol. Chem., 2000, 275, 40810–40816 CrossRef CAS.
- M. Chevillard-Briet, D. Trouche and L. Vandel, Control of CBP co-activating activity by arginine methylation, EMBO J., 2002, 21, 5457–5466 CrossRef CAS.
- S. Pal, S. N. Vishwanath, H. Erdjument-Bromage, P. Tempst and S. Sif, Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes, Mol. Cell. Biol., 2004, 24, 9630–9645 CrossRef CAS.
- V. Verbiest, D. Montaudon, M. T. Tautu, J. Moukarzel and J. P. Portail, et al., Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins, FEBS Lett., 2008, 582, 1483–1489 CrossRef CAS.
- W. J. Lin, J. D. Gary, M. C. Yang, S. Clarke and H. R. Herschman, The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase, J. Biol. Chem., 1996, 271, 15034–15044 CrossRef CAS.
- K. Higashimoto, P. Kuhn, D. Desai, X. Cheng and W. Xu, Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12318–12323 CrossRef CAS.
- Q. Feng, B. He, S. Y. Jung, Y. Song and J. Qin, et al., Biochemical control of CARM1 enzymatic activity by phosphorylation, J. Biol. Chem., 2009, 284, 36167–36174 CrossRef CAS.
- U. Dery, Y. Coulombe, A. Rodrigue, A. Stasiak and S. Richard, et al., A glycine-arginine domain in control of the human MRE11 DNA repair protein, Mol. Cell. Biol., 2008, 28, 3058–3069 CrossRef CAS.
- F. M. Boisvert, U. Dery, J. Y. Masson and S. Richard, Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control, Genes Dev., 2005, 19, 671–676 CrossRef CAS.
- D. Cheng, N. Yadav, R. W. King, M. S. Swanson and E. J. Weinstein, et al., Small molecule regulators of protein arginine methyltransferases, J. Biol. Chem., 2004, 279, 23892–23899 CrossRef CAS.
- S. L. McGovern, E. Caselli, N. Grigorieff and B. K. Shoichet, A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening, J. Med. Chem., 2002, 45, 1712–1722 CrossRef CAS.
- J. Trapp, R. Meier, D. Hongwiset, M. U. Kassack and W. Sippl, et al., Structure–activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins), ChemMedChem, 2007, 2, 1419–1431 CrossRef CAS.
- R. Ragno, S. Simeoni, S. Castellano, C. Vicidomini and A. Mai, et al., Small molecule inhibitors of histone arginine methyltransferases: homology modeling, molecular docking, binding mode analysis, and biological evaluations, J. Med. Chem., 2007, 50, 1241–1253 CrossRef CAS.
- A. Mai, D. Cheng, M. T. Bedford, S. Valente and A. Nebbioso, et al., epigenetic multiple ligands: mixed histone/protein methyltransferase, acetyltransferase, and class III deacetylase (sirtuin) inhibitors, J. Med. Chem., 2008, 51, 2279–2290 CrossRef CAS.
- K. Balasubramanyam, R. A. Varier, M. Altaf, V. Swaminathan and N. B. Siddappa, et al., Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription, J. Biol. Chem., 2004, 279, 51163–51171 CrossRef CAS.
- J. Trapp, A. Jochum, R. Meier, L. Saunders and B. Marshall, et al., Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition, J. Med. Chem., 2006, 49, 7307–7316 CrossRef CAS.
- K. Bonham; S. Hemmers; Y. H. Lim; D. M. Hill; M. G. Finnet al., Effects of a novel arginine methyltransferase inhibitor on T-helper cell cytokine production. Febs J, 277, pp. 2096–2108 Search PubMed.
- F. Blanchet, A. Cardona, F. A. Letimier, M. S. Hershfield and O. Acuto, CD28 costimulatory signal induces protein arginine methylation in T cells, J. Exp. Med., 2005, 202, 371–377 CrossRef CAS.
- S. Richard, M. Morel and P. Cleroux, Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5), Biochem. J., 2005, 388, 379–386 CrossRef CAS.
- K. A. Mowen, B. T. Schurter, J. W. Fathman, M. David and L. H. Glimcher, Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes, Mol. Cell, 2004, 15, 559–571 CrossRef CAS.
- A. V. Purandare, Z. Chen, T. Huynh, S. Pang and J. Geng, et al., Pyrazole inhibitors of coactivator associated arginine methyltransferase 1 (CARM1), Bioorg. Med. Chem. Lett., 2008, 18, 4438–4441 CrossRef CAS.
- A. Spannhoff, R. Heinke, I. Bauer, P. Trojer and E. Metzger, et al., Target-based approach to inhibitors of histone arginine methyltransferases, J. Med. Chem., 2007, 50, 2319–2325 CrossRef CAS.
- A. Spannhoff, R. Machmur, R. Heinke, P. Trojer and I. Bauer, et al., A novel arginine methyltransferase inhibitor with cellular activity, Bioorg. Med. Chem. Lett., 2007, 17, 4150–4153 CrossRef CAS.
- R. Heinke, A. Spannhoff, R. Meier, P. Trojer and I. Bauer, et al., Virtual screening and biological characterization of novel histone arginine methyltransferase PRMT1 inhibitors, ChemMedChem, 2009, 4, 69–77 CrossRef CAS.
- H. Wu; J. Min; V. V. Lunin; T. Antoshenko; L. Dombrovskiet al., Structural biology of human H3K9 methyltransferases. PLoS One, 5, p. e8570 Search PubMed.
- T. J. Wigle; J. M. Herold; G. A. Senisterra; M. Vedadi; D. B. Kireevet al., Screening for inhibitors of low-affinity epigenetic peptide-protein interactions: an AlphaScreen-based assay for antagonists of methyl-lysine binding proteins. J Biomol Screen, 15, pp. 62–71 Search PubMed.
- A. M. Quinn; M. T. Bedford; A. Espejo; A. Spannhoff; C. P. Austinet al., A homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics. Nucleic Acids Res, 38, p. e11 Search PubMed.
- M. Y. Kang, B. B. Lee, Y. H. Kim, D. K. Chang and S. Kyu Park, et al., Association of the SUV39H1 histone methyltransferase with the DNA methyltransferase 1 at mRNA expression level in primary colorectal cancer, Int. J. Cancer, 2007, 121, 2192–2197 CrossRef CAS.
- K. M. McGarvey, J. A. Fahrner, E. Greene, J. Martens and T. Jenuwein, et al., Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state, Cancer Res., 2006, 66, 3541–3549 CrossRef CAS.
- F. Aniello, G. Colella, G. Muscariello, A. Lanza and D. Ferrara, et al., Expression of four histone lysine-methyltransferases in parotid gland tumors, Anticancer Res., 2006, 26, 2063–2067 CAS.
- K. Saigo, K. Yoshida, R. Ikeda, Y. Sakamoto and Y. Murakami, et al., Integration of hepatitis B virus DNA into the myeloid/lymphoid or mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in human hepatocellular carcinoma, Hum. Mutat., 2008, 29, 703–708 CrossRef CAS.
- I. M. Bachmann, O. J. Halvorsen, K. Collett, I. M. Stefansson and O. Straume, et al., EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast, J. Clin. Oncol., 2005, 24, 268–273 CrossRef.
- A. E. McBride, J. T. Cook, E. A. Stemmler, K. L. Rutledge and K. A. McGrath, et al., Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions, J. Biol. Chem., 2005, 280, 30888–30898 CrossRef CAS.
- R. Meyer, S. S. Wolf and M. Obendorf, PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor, J. Steroid Biochem. Mol. Biol., 2007, 107, 1–14 CrossRef CAS.
- S. El Messaoudi, E. Fabbrizio, C. Rodriguez, P. Chuchana and L. Fauquier, et al., Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 13351–13356 CrossRef CAS.
- H. Hong, C. Kao, M. H. Jeng, J. N. Eble and M. O. Koch, et al., Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status, Cancer, 2004, 101, 83–89 CrossRef CAS.
- Y. H. Lee, S. A. Coonrod, W. L. Kraus, M. A. Jelinek and M. R. Stallcup, Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 3611–3616 CrossRef CAS.
- S. Pal, R. A. Baiocchi, J. C. Byrd, M. R. Grever and S. T. Jacob, et al., Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma, EMBO J., 2007, 26, 3558–3569 CrossRef CAS.
- X. Le Guezennec, M. Vermeulen, A. B. Brinkman, W. A. Hoeijmakers and A. Cohen, et al., MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties, Mol. Cell. Biol., 2006, 26, 843–851 CrossRef CAS.
- C. P. Tan and S. Nakielny, Control of the DNA methylation system component MBD2 by protein arginine methylation, Mol. Cell. Biol., 2006, 26, 7224–7235 CrossRef CAS.
- D. Hyllus, C. Stein, K. Schnabel, E. Schiltz and A. Imhof, et al., PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation, Genes Dev., 2007, 21, 3369–3380 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2010 |
