Bioluminescent nanosensors for protease detection based upon gold nanoparticle–luciferase conjugates†
Young-Pil
Kim
a,
Weston L.
Daniel
b,
Zuyong
Xia
a,
Hexin
Xie
a,
Chad A.
Mirkin
b and
Jianghong
Rao
*a
aMolecular Imaging Program at Stanford (MIPS), Departments of Radiology and Chemistry, Stanford University, Stanford, CA 94305-5484, USA. E-mail: jrao@stanford.edu; Fax: +1 650 736 7925; Tel: +1 650 736 8563
bDepartment of Chemistry and International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA. E-mail: chadnano@northwestern.edu; Fax: +1 847 467 5123; Fax: +1 847 467 2907
First published on 12th October 2009
Abstract
This communication reports the use of click chemistry to site-specifically conjugate bioluminescent Renilla luciferase proteins to gold nanoparticles (Au NPs) for sensing protease activity. The bioluminescent emission from luciferase was efficiently quenched by Au NPs, but significantly recovered after the proteolytic cleavage.
Nanoparticles functionalized with biomolecules hold promise for many potential applications in nanotechnology and nanomedicine.1–7 Among the many types of nanoparticles, gold nanoparticles (Au NPs) have been of special interest due to their biocompatibility and outstanding biophysical properties and have already been utilized in many medical applications.8–10 It has been demonstrated that Au NPs have superior quenching efficiency compared to molecular quenchers for emission from small organic dyes,11 fluorescent proteins,12 and even semiconductor quantum dots.13 This quenching ability enables the design of fluorescent nanosensors based on Au NPs for biosensing applications.14–17 However, these fluorescent probes are often amenable to fast photobleaching and excitation-induced cytotoxicity.18 Bioluminescence (BL) offers an alternative to circumvent this problem and has been drawing much attention for in vitro monitoring19 and noninvasive imaging of specific targets in vivo.20 While small organic dyes have been reported to quench bioluminescence emission through bioluminescence resonance energy transfer,21 the utility of Au NPs as luminescence quenchers has not been explored in the literature. Herein, we show that Au NPs can efficiently quench the emission from the bioluminescent protein luciferase and that this Au NP quenching effect can be applied to design nanosensors for the detection of matrix metalloproteinase-2 (MMP-2) activity.
The bioluminescent protein used in this work is a Renilla reniformis luciferase mutant dubbed Luc8, which has eight mutations and shows higher stability and improved catalytic efficiency than the wild-type luciferase.22 A site-specific bioconjugation strategy was devised to conjugate the Luc8 to Au NPs with click chemistry and an intein-mediated protein splicing reaction (Scheme 1). The cycloaddition between azides and alkynes, known as the “click” reaction, is highly efficient and specific, can take place in water, and has been successfully applied to in vitro and in vivo applications, including the coupling of biomolecules to nanoparticles.23–26 An intein-mediated ligation method27,28 was implemented to site-specifically introduce the azide moiety to Luc8 to facilitate the conjugation. MMP-2 was chosen as the model protease because of its important role in promoting tumor progression and invasion.29,30 A single peptide substrate of MMP-2 (IPVSLRSG) was employed as the sensing domain in the nanoconjugate.
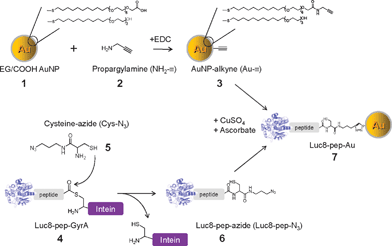 | ||
| Scheme 1 Conjugation of gold nanoparticles (Au NPs) with recombinant luciferase proteins via click chemistry and an intein-based ligation method. | ||
As depicted in Scheme 1, oligo(ethylene glycol)-modified Au NPs 1 (∼5 nm Au core diameter) with carboxyl groups at their termini were modified with an alkyne derivative 2via a carbodiimide-mediated reaction, resulting in alkynated Au NP (3, Au–![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) ). A recombinant Luc8 protein (4, Luc8-pep-GyrA) was expressed with the substrate peptide (IPVSLRSG) and the Mex GyrA intein protein fused at its C-terminus.27 The thioester intermediate, formed via the catalysis of GyrA, was reacted with cysteine-azide (5, Cys-N3) to produce the Luc8-pep fusion with an azide-modified C-terminus (6, Luc8-pep-N3). The final product (7, Luc8-pep-Au NP) was generated from 3 and 6 by copper(I)-catalyzed azide–alkyne [3 + 2] cycloaddition.
). A recombinant Luc8 protein (4, Luc8-pep-GyrA) was expressed with the substrate peptide (IPVSLRSG) and the Mex GyrA intein protein fused at its C-terminus.27 The thioester intermediate, formed via the catalysis of GyrA, was reacted with cysteine-azide (5, Cys-N3) to produce the Luc8-pep fusion with an azide-modified C-terminus (6, Luc8-pep-N3). The final product (7, Luc8-pep-Au NP) was generated from 3 and 6 by copper(I)-catalyzed azide–alkyne [3 + 2] cycloaddition.
We examined the relative migration of the Luc8-pep-GyrA protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to confirm its intein-based ligand modification by 5 (Cys-N3). Luc8-pep-GyrA showed two strong bands under non-reducing conditions (lane 1 in Fig. 1) corresponding to the monomeric form (59 kDa) and the oxidized dimer form (118 kDa) of the protein. Addition of only the Cys-N3 group to the fusion protein resulted in few cleavage products (lane 2). However, efficient cleavage by Cys-N3 took place in the presence of 2-mercaptoethanesulfonic acid (MESA) as evidenced by the reduced intensity of fusion protein band, and the appearance of two bands corresponding to the azide-modified Luc8 fusion protein and the free GyrA intein (lane 4). In comparison, MESA treatment alone also produced two cleavage bands (lane 3) corresponding to the Luc8-pep (37 kDa) and the GyrA intein (21 kDa), but the unmodified Luc8-pep appeared to have a slightly faster mobility than the azide-modified Luc8-pep. Purification with a Ni-nitrilotriacetic acid (NTA) affinity column removed the His6 tagged GyrA intein and any uncleaved fusion protein from the reaction and gave the pure azide-modified fusion protein (Luc8-pep-N3) (lane 5). This result confirmed the successful site-specific introduction of the azide group to the Luc8 fusion protein.
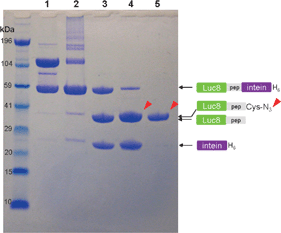 | ||
| Fig. 1 NuPAGE analysis of cysteine-N3 ligation by intein-mediated cleavage. Luc8-pep-GyrA protein (lane 1) was incubated with different thiol groups for 16 h at 4 °C: 10 mM Cys-N3 (lane 2), 20 mM MESA (lane 3), and 20 mM MESA/10 mM Cys-N3 (lane 4). The purified sample from lane 4 is displayed in lane 5. The reaction mixture was loaded into each well after mixing with lithium dodecyl sulfate (LDS) loading buffer in the absence of reducing agent. The arrows indicate the position of Luc8 protein fused with the Cys-N3. The molecular weight standard is displayed in the far left lane. Electrophoresis was carried out on 12% gel in bis (2-hydroxyethyl)amino-tris(hydroxymethyl)methane (Bis-Tris) buffer containing LDS for 45 min. | ||
Au–![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) was treated with Luc8-pep-N3 in the presence of CuSO4 and ascorbate to couple the Au NPs with azide-modified luciferase by click chemistry. The conjugation of the protein to the Au NP surface was confirmed with electrophoresis, UV-Vis spectroscopy, and a Bradford assay. The Luc8 conjugated Au NPs displayed a strong band shift in agarose gel electrophoresis (lane 2 in Fig. 2(a)) when compared to the negative controls lacking either Luc8-pep-N3 (lane 1), CuSO4 (lane 3), or ascorbate (lane 4). This result clearly indicated that the Cu(I)-mediated click reaction enabled the conjugation of Au–
was treated with Luc8-pep-N3 in the presence of CuSO4 and ascorbate to couple the Au NPs with azide-modified luciferase by click chemistry. The conjugation of the protein to the Au NP surface was confirmed with electrophoresis, UV-Vis spectroscopy, and a Bradford assay. The Luc8 conjugated Au NPs displayed a strong band shift in agarose gel electrophoresis (lane 2 in Fig. 2(a)) when compared to the negative controls lacking either Luc8-pep-N3 (lane 1), CuSO4 (lane 3), or ascorbate (lane 4). This result clearly indicated that the Cu(I)-mediated click reaction enabled the conjugation of Au–![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) to Luc8-pep-N3.31 We further confirmed this conjugation using UV-Vis spectroscopy after purifying the mixture with 100 kDa cut-off microfiltration. Compared to the unconjugated Au NPs (Au–
to Luc8-pep-N3.31 We further confirmed this conjugation using UV-Vis spectroscopy after purifying the mixture with 100 kDa cut-off microfiltration. Compared to the unconjugated Au NPs (Au–![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) ), which had a strong absorption at 516 nm, the Luc8-conjugated Au NPs displayed a slightly red-shifted spectrum (520 nm) because of their surface modification (Fig. 2(b)). Finally, we determined that the conjugation afforded 4.7 ± 1.2 proteins per Au NP by displacing its monolayer with 2-mercaptoethanol and quantifying the luciferase concentration with a Bradford assay (ESI,† Fig. S1).
), which had a strong absorption at 516 nm, the Luc8-conjugated Au NPs displayed a slightly red-shifted spectrum (520 nm) because of their surface modification (Fig. 2(b)). Finally, we determined that the conjugation afforded 4.7 ± 1.2 proteins per Au NP by displacing its monolayer with 2-mercaptoethanol and quantifying the luciferase concentration with a Bradford assay (ESI,† Fig. S1).
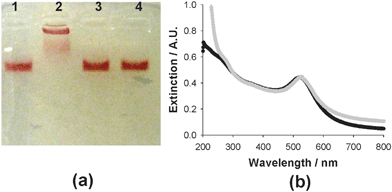 | ||
Fig. 2 Click chemistry-based conjugation of Au–![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) and Luc8-pep-N3: agarose (1.2%) gel electrophoresis (a) and UV-visible spectroscopy analysis (b). In agarose gel electrophoresis, Au– and Luc8-pep-N3: agarose (1.2%) gel electrophoresis (a) and UV-visible spectroscopy analysis (b). In agarose gel electrophoresis, Au–![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) was subjected to different reaction conditions, where one of the components was removed in the reaction solution: without Luc8-pep-N3 (lane 1), without CuSO4 (lane 3), or without ascorbate (lane 4). The extinction graphs were displayed for Au– was subjected to different reaction conditions, where one of the components was removed in the reaction solution: without Luc8-pep-N3 (lane 1), without CuSO4 (lane 3), or without ascorbate (lane 4). The extinction graphs were displayed for Au–![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) (black line) and for Luc8-pep-Au NP (gray line). (black line) and for Luc8-pep-Au NP (gray line). | ||
Before using the Luc8-pep-Au NP conjugates in the MMP-2 detection assay, we examined the effect of Cu(I) on the activity of the Luc8 protein, because it has been reported to deactivate the function of the protein.32 When the Luc8 was treated with Cu(II) and ascorbate the activity of the Luc8 decreased when compared to the untreated protein. However, this activity loss could be partially recovered by treatment of the protein with L-cysteine. Up to 40% of the initial BL signal of Luc8 was regained (ESI,† Fig. S2). Among the other Cu(I)–chelating agents that were tested, dithiothreitol (DTT) was also effective at restoring the Luc8 activity. However, DTT displaced the oligo(ethylene glycol) groups from the Au NP surface33 and destabilized the Au NPs. To further verify the quenching efficiency of Au NP, the carboxyl Au NP was directly conjugated with Luc8-pep-N3via 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) reaction with different ratios of Luc8 to Au NP, and the maximum quenching efficiency was observed at the ratios of 2 : 1 and 1 : 1 (Luc8 : Au NP), which corresponds to 85%, as compared to the initial BL (ESI,† Fig. S3).
Finally, we evaluated whether the quenched bioluminescent nanosensor could measure the activity of MMP-2, which specifically cleaves the amide bond between Ser-Lys34 in the peptide sequence linking the Luc8 to the Au NP. The BL emission of the Luc8-pep-Au NP conjugate solution was enhanced about 15-fold over the background signal 1 h after adding MMP-2 (Fig. 3(a)). Sufficient enzymatic cleavage was observed within 1 h under the given conditions (ESI,† Fig. S4), and the increase in BL signal was linearly dependent on the logarithmic concentration of MMP-2, with a dynamic range from 50 ng mL−1 to 1 μg mL−1 (Fig. 3(b)). As a control, a Luc8-pep-Au NP nanoconjugate was prepared via an EDC-mediated coupling reaction between the lysine residues of the Luc8-pep-N3 and the carboxyl groups of the Au NP. As expected, this conjugate did not show a MMP-2 dependent increase in bioluminescence over the background intensity (Fig. 3(c) and (d)). This result indicates that the site-specific orientation of Luc8 on the Au NP is critical for detecting protease activity.
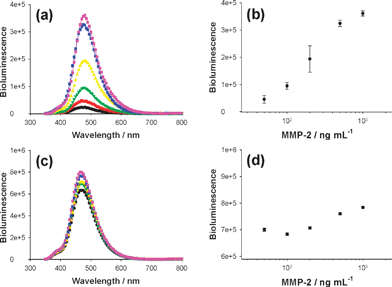 | ||
| Fig. 3 Detection of protease activity by using different Luc8-pep-Au NP conjugates: click and intein-based conjugate (a and b) and randomly coupled conjugate (c and d). Left graphs represent bioluminescence signals with different MMP-2 enzyme concentrations (0, 50, 100, 200, 500, and 1000 ng mL−1, from bottom to top), and the correlation between peak intensity and enzyme concentration is displayed in right graphs. | ||
In conclusion, we demonstrated the conjugation of luciferase to Au NPs via click chemistry and an intein-mediated ligation and the subsequent detection of protease activity based on the Au NP-quenched BL. The Cu(I)-catalyzed click reaction allowed for a rapid and efficient conjugation between the nanoparticle and the luciferase protein, and the intein-mediated ligation enabled this protein to be site-specifically immobilized on the Au NP, both of which are critical for the function of the nanosensor. This conjugation method should be generally applicable to other proteins and nanoparticles. Moreover, the Au NP–luciferase system can avoid the disadvantages in fluorescence-based approaches such as photoexcitation-derived photobleaching and phototoxicity. We anticipate that our method will help expand the utility of functionalized nanoparticles in biology and nanotechnology research.
This work was supported in part by the National Cancer Institute (Grant 1R01CA135294-01), the Burroughs Wellcome Fund, the Stanford University National Cancer Institute Centers of Cancer Nanotechnology Excellence (Grant 1U54CA119367-01), the Northwestern University National Cancer Institute Centers of Cancer Nanotechnology Excellence, and the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2009-352-D00098). C. A. M. is also grateful for a National Security Science and Engineering Faculty Fellows Program award.
Notes and references
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607 CrossRef CAS
.
- N. L. Rosi and C. A. Mirkin, Chem. Rev., 2005, 105, 1547 CrossRef CAS
.
- M. Bruchez, Jr, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Science, 1998, 281, 2013 CrossRef CAS
.
- E. Katz and I. Willner, Angew. Chem., Int. Ed., 2004, 43, 6042 CrossRef CAS
.
- M. Ferrari, Nat. Rev. Cancer, 2005, 5, 161 CrossRef CAS
.
- C. M. Niemeyer, B. Ceyhan and P. Hazarika, Angew. Chem., Int. Ed., 2003, 42, 5766 CrossRef CAS
.
- R. Elghanian, J. J. Storhoff, R. C. Mucic, R. L. Letsinger and C. A. Mirkin, Science, 1997, 277, 1078 CrossRef CAS
.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. Lytton-Jean, M. S. Han and C. A. Mirkin, Science, 2006, 312, 1027 CrossRef CAS
.
- J. M. Nam, C. S. Thaxton and C. A. Mirkin, Science, 2003, 301, 1884 CrossRef CAS
.
- R. A. Sperling, P. Rivera Gil, F. Zhang, M. Zanella and W. J. Parak, Chem. Soc. Rev., 2008, 37, 1896 RSC
.
- B. Dubertret, M. Calame and A. J. Libchaber, Nat. Biotechnol., 2001, 19, 365 CrossRef CAS
.
- P. Hazarika, F. Kukolka and C. M. Niemeyer, Angew. Chem., Int. Ed., 2006, 45, 6827 CrossRef CAS
.
- Y. P. Kim, Y. H. Oh, E. Oh, S. Ko, M. K. Han and H. S. Kim, Anal. Chem., 2008, 80, 4634 CrossRef CAS
.
- A. Verma, H. Nakade, J. M. Simard and V. M. Rotello, J. Am. Chem. Soc., 2004, 126, 10806 CrossRef CAS
.
- D. J. Maxwell, J. R. Taylor and S. Nie, J. Am. Chem. Soc., 2002, 124, 9606 CrossRef CAS
.
- E. Oh, M. Y. Hong, D. Lee, S. H. Nam, H. C. Yoon and H. S. Kim, J. Am. Chem. Soc., 2005, 127, 3270 CrossRef CAS
.
- D. S. Seferos, D. A. Giljohann, H. D. Hill, A. E. Prigodich and C. A. Mirkin, J. Am. Chem. Soc., 2007, 129, 15477 CrossRef CAS
.
- R. A. Hoebe, C. H. Van Oven, T. W. Gadella, Jr, P. B. Dhonukshe, C. J. Van Noorden and E. M. Manders, Nat. Biotechnol., 2007, 25, 249 CrossRef CAS
.
- Y. Zhang, M. K. So, A. M. Loening, H. Yao, S. S. Gambhir and J. Rao, Angew. Chem., Int. Ed., 2006, 45, 4936 CrossRef CAS
.
- P. R. Contag, I. N. Olomu, D. K. Stevenson and C. H. Contag, Nat. Med., 1998, 4, 245 CrossRef CAS
.
- Z. Xia and J. Rao, Curr. Opin. Biotechnol., 2009, 20, 37 CrossRef CAS
.
- A. M. Loening, T. D. Fenn, A. M. Wu and S. S. Gambhir, Protein Eng., Des. Sel., 2006, 19, 391 CrossRef CAS
.
- H. C. Kolb and K. B. Sharpless, Drug Discovery Today, 2003, 8, 1128 CrossRef CAS
.
- C. R. Becer, R. Hoogenboom and U. S. Schubert, Angew. Chem., Int. Ed., 2009, 48, 4900 CrossRef CAS
.
- J. L. Brennan, N. S. Hatzakis, T. R. Tshikhudo, N. Dirvianskyte, V. Razumas, S. Patkar, J. Vind, A. Svendsen, R. J. Nolte, A. E. Rowan and M. Brust, Bioconjugate Chem., 2006, 17, 1373 CrossRef CAS
.
- G. von Maltzahn, Y. Ren, J. H. Park, D. H. Min, V. R. Kotamraju, J. Jayakumar, V. Fogal, M. J. Sailor, E. Ruoslahti and S. N. Bhatia, Bioconjugate Chem., 2008, 19, 1570 CrossRef CAS
.
- Z. Xia, Y. Xing, M. K. So, A. L. Koh, R. Sinclair and J. Rao, Anal. Chem., 2008, 80, 8649 CrossRef CAS
.
- P. C. Lin, S. H. Ueng, M. C. Tseng, J. L. Ko, K. T. Huang, S. C. Yu, A. K. Adak, Y. J. Chen and C. C. Lin, Angew. Chem., Int. Ed., 2006, 45, 4286 CrossRef CAS
.
- A. John and G. Tuszynski, Pathol. Oncol. Res., 2001, 7, 14 Search PubMed
.
- M. Egeblad and Z. Werb, Nat. Rev. Cancer, 2002, 2, 163 CrossRef
.
- The combination of Au–
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) and Luc8-pep-N3 had a much greater yield compared to when the azide was on the Au NP and alkyne was on the Luc8 under the same reaction conditions.
and Luc8-pep-N3 had a much greater yield compared to when the azide was on the Au NP and alkyne was on the Luc8 under the same reaction conditions. - A. J. Link and D. A. Tirrell, J. Am. Chem. Soc., 2003, 125, 11164 CrossRef CAS
.
- C. S. Thaxton, H. D. Hill, D. G. Georganopoulou, S. I. Stoeva and C. A. Mirkin, Anal. Chem., 2005, 77, 8174 CrossRef CAS
.
- B. E. Turk, L. L. Huang, E. T. Piro and L. C. Cantley, Nat. Biotechnol., 2001, 19, 661 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, the determination of the number of Luc8 per Au NP, signal recovery of Luc8 after the click reaction, the quenching efficiency of the conjugate, and time-dependent monitoring of MMP-2 activity. See DOI: 10.1039/b915612g |
| This journal is © The Royal Society of Chemistry 2010 |
