Template-controlled conformational patterns of insulin fibrillar self-assembly reflect history of solvation of the amyloid nuclei
Wojciech
Dzwolak
*a,
Ralf
Jansen
b,
Vytautas
Smirnovas
ab,
Anna
Loksztejn
a,
Sylwester
Porowski
a and
Roland
Winter
*b
aInstitute of High Pressure Physics, Polish Academy of Sciences, Sokolowska 29/37, 01-142 Warsaw, Poland
bUniversity of Dortmund, Department of Chemistry, Physical Chemistry I-Biophysical Chemistry, Otto-Hahn Str. 6, D-44227 Dortmund, Germany
First published on 28th February 2005
Abstract
In the presence of ethanol, insulin forms amyloid morphologically distinct from the ambient specimen. Due to stability of fibrils and the autocatalytic character of the process, the two amyloid templates, when seeded, replicate the initial morphologies (and inter-β-strand hydrogen bonding patterns) regardless of the environmental biases, such as the cosolvent presence. Such “templated memory” effect is advantageous in synthesizing structurally uniform protein nanofibrils under conditions favoring alternative “wild” forms. This also appears to parallel “prion strains” phenomenon, suggesting that “strains” may reflect a generic trait in all amyloids including those not associated with disease.
The amyloidogenic self-assembly of proteins, which results in formation of β-sheet rich fibrils, has been implicated in a number of neurodegenerative diseases, such as Alzheimer’s disease, or (the prion-associated) Creutzfeldt–Jakob disease.1 Although the mechanisms of protein aggregation remain still largely unclear, the phenomenon has already been exploited for the sake of possible nanotechnological applications, including new biomaterials,2 conducting nanowires,3etc. Amyloid formation is a profoundly irreversible and kinetically controlled process, which may be triggered by genomic (destabilizing mutations), or non-genomic (denaturants, low pH) factors. Amyloid preparations are often morphologically heterogeneous,4 which hampers detailed structural studies and nanotechnology-oriented applications. Since perturbation of hydrational forces affects stability of insulin aggregation nuclei and leads to distinct fibrillar types,5,6 we were seeking whether such solvent-programmed conformations may act as templates inducing conserved fibrillar types, even under conditions favoring different morphologies. Here we show that transient ethanol-induced perturbation of hydration of insulin aggregation nuclei results in a conformational variant of the ambient amyloid with distinct morphological traits. According to earlier studies, aggregation of insulin is an energetically self-sustaining process7 that permits seeding the amyloid at room temperature. Cosolvents enhance fibrils distinct both in terms of morphology and hydrogen-bonding patterns within aggregated β-strands, as seen in FT-IR spectroscopy.5,6 Such patterns, when induced through diluted ethanol, are preserved upon cross-seeding to aqueous solutions of the native insulin.8
A morphological probe of the homogenous and heterogonous seeding of native insulin with its “ambient”, and conformational variant fibrils has been carried out with high-resolution atomic force microscopy (AFM). Fig. 1 presents images of the fibrils formed at 25 °C by seeding of dimeric insulin (in water, 1.8 pH), or insulin monomers (in 20 wt.% EtOH, pH 1.8) with sonificated fibrils pre-formed spontaneously through an incubation of unstirred protein solutions for 24 h at 60 °C. The fibrils formed at 60 °C in water are long, straight and unbranched. In the presence of 20 wt.% EtOH, thinner, and curved forms prevail. Once formed, either type is stable and does not change its outlook upon transferring to an alternative environment. Seeding of unstirred protein solutions (either in water or in 20 wt.% ethanol) was carried out for 24 h at 25 °C. The images of daughter fibrils in Fig. 1 convey a clear proof that the homogenously seeded amyloids (b,c), i.e. under solvent conditions corresponding to preparation of the zero generation fibrils, retain the original templates’ morphologies. Interestingly, according to the cross-seeding data (a,d), the type of template, not the solvent conditions of seeding, determine the morphology of the subsequent daughter generations of fibrils. This parallels the cross-seeding FT-IR spectroscopy data, suggesting that the two different morphologies are a more tangible aspect of the existence of two distinct conformational variants of insulin amyloid, both of which possess strong autocatalytic properties enabling “hijacking” of thermodynamically favorable self-assembly pathways.8 The position of the amide I′ band confirms that β-sheet is the basic secondary structural motif of either fibrillar type. However, the exact position of the band depends again on the presence of ethanol in the case of spontaneous amyloidogenesis at 60 °C (the zero generation fibrils in Fig. 2A and 2B), or the history of the template in the case of seeded amyloidogenesis at 25 °C. In the second-derivative spectra (Fig. 2A), this is reflected by the dramatic reversal of the relative intensities of the two spectral components around 1633 and 1620 cm−1 assigned to two populations of intra-fibrillar β-strands with different hydrogen bonding energies. The effect of the spectral pattern-defining template persists at least for two consecutive generations of cross-seeded fibrils (Fig. 2A). Further experiments with homogeneous-following-heterogeneous (and vice versa) seedings, summarized in Fig. 2B, support the notion that the original, unseeded templates determine the spectral features of the following generations of seeded fibrils, regardless of environment biases. This observation is in accordance with the previously discussed AFM data.
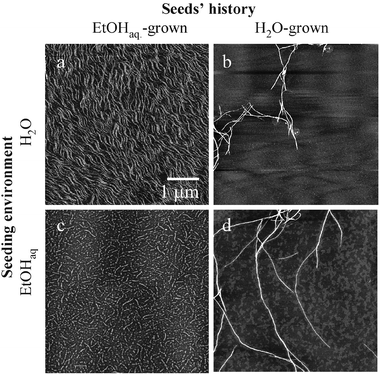 | ||
| Fig. 1 AFM images of the first generation of insulin amyloid fibrils seeded homogeneously (b,c) and cross-seeded (a,d) reflect prevalence of the template’s self-assembly history over the environment’s properties. The curvy, and scattered insulin amyloid formed spontaneously in the presence of ethanol reproduces the same morphology whether seeded homogenously to insulin in 20% EtOHaq (c), or cross-seeded to water-dissolved protein (a). Likewise, the long, straight fibrils from aqueous amyloid preparations proliferate maintaining the original morphology either in the homogeneous- (b) or cross-seeding (d). The initial templates (“zero” generation of fibrils) were formed through a 60 °C/24 h incubation of 2 wt.% bovine insulin (Sigma, USA) in H2O, pH 1.8; or in 20 wt.% EtOHaq, pH 1.8. Seeding was carried out at 25 °C, through a 24 h incubation of native insulin (2 wt.% in H2O, pH 1.8, or in 20 wt.% ethanol, pH 1.8) doped with sonificated “zero” generation amyloids at a 20∶1 wt. ratio. The amyloid samples were diluted with H2O 400 times prior to deposition on mica. All AFM images were recorded in the tapping-in-air height mode on a MultiMode™ SPM microscope with a Nanoscope IIIa Controller (Digital Instruments, USA). The 1 μm scale bar applies to all the four (a,b,c,d) images. | ||
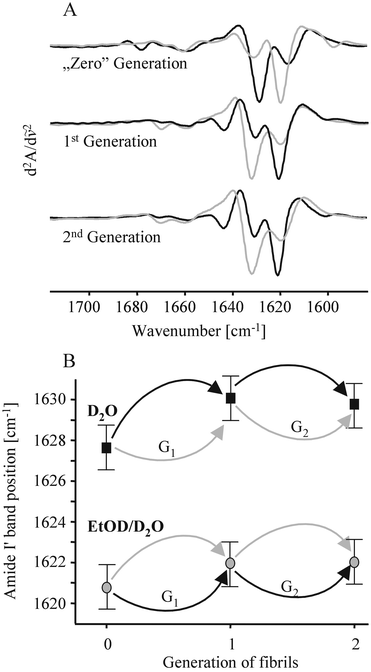 | ||
| Fig. 2 (A) Second derivative FT-IR spectra of the amide I′ band of the zeroth, first and second generations of cross-seeded insulin amyloid. Each spectrum’s shade marks the type of insulin solution in which fibrils were formed: aqueous is black, and EtODaq is gray. Sample preparation regimes were similar to the AFM experiment, except that all compounds were deuterated, and samples were not diluted prior to spectral data acquisition. The second-generation spectra correspond to samples, where the original “zero generation” templates were formed under conditions opposite to each of the two consecutive cross-seeding. (B) The final amide I′ band’s peak position of the zeroth, first and second generations of cross-seeded insulin amyloid. The arrows denote conditions of seeding: D2O (black), 20 wt.% EtODaq (gray). The seeding pathways leading to the first and second generations of fibrils from Fig. 2A are labeled as G1, and G2, respectively. | ||
The seeding patterns of insulin amyloid reported herein could be concluded to be mirroring a “templated memory” of the protein fibrils, which stores solvational history of the original seeds, but not properties of the environments, in which growth of following seeded generations continued (Fig. 3). An actual nucleation event (i.e., stacking of destabilized insulin molecules into an amyloid template) takes place only upon the unseeded amyloidogenesis at 60 °C.9 The temperature-induced unfolding of insulin causes exposure of hydrophobic side-chains and renders the protein molecules sensitive to forces of EtOH-controlled preferential hydration and binding.10 During the seeding at 25 °C, nucleation is kinetically marginalized by template-assisted elongation of fibrils. In other words, a thermodynamic control (through hydrational/solvational forces) of insulin self-assembly patterns, is feasible only during the nucleation step, which occurs in the lag phase of a spontaneous aggregation. Once the nucleus is formed and the elongation of a fibril sets in, the now kinetically controlled process tends to ignore environmental biases. Subtle solvational effects have been implicated in dramatic changes of the aggregation propensity of model polypeptides.11
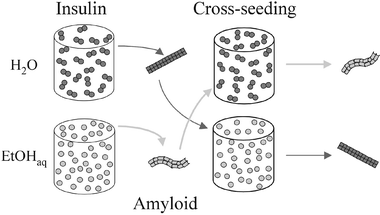 | ||
| Fig. 3 The schematic summary of the insulin cross-seeding experiments shows how a “foreign” template is overriding a thermodynamically preferred (under particular cosolvent conditions) aggregation pathway and reproduces its own morphology. | ||
The presented case of fibril morphology controlled by the solvational history of aggregation nuclei opens up new prospects for the in situ synthesis of biomaterials, such as scaffolds that override unfavorable transient solvent conditions and maintain the “programmed” topology. At the same time, the fact that two morphologically-distinguishable amyloidogenic templates may be induced in a single, covalently unmodified amino acid sequence parallels the prion strains phenomenon, and by the same token is an argument for a generic character of the strains.12 In fact, a very recent study reported similar behavior of two self propagating templates obtained from the Aβ1–40 Alzheimer peptide.13 The authors employed mechanical agitation of the peptide solution as the external perturbation. Perhaps, the cosolvent-effect used herein may be explored in a more comprehensive way in terms of preferential hydration and binding, and therefore offer means to explain physicochemical mechanisms leading to occurrence of different amyloid strains.
Acknowledgements
RW acknowledges financial support from the Deutsche Forschungsgemeinschaft (DFG-FOR 436). WD acknowledges financial support from the State Committee for Scientific Research, Grant No. 2P04A 011 28 (2005–2007). WD and RW also acknowledge support from the EU COST D30 action “High Pressure Tuning of Biochemical Processes: Protein Dynamics and Aggregation”.References
- C. M. Dobson, Trends Biochem. Sci., 1999, 24, 329 CrossRef CAS.
- S. G. Zhang, Nat. Biotechnol., 2003, 21, 1171 CrossRef CAS.
- Scheibel, R. Parthasarathy, G. Sawicki, X. M. Lin, H. Jaeger and S. L. Lindquist, Proc. Natl. Acad. Sci. USA, 2003, 100, 4527 CrossRef CAS.
- R. Jansen, W. Dzwolak and R. Winter, Biophys. J., 2005, 88, 1344 CAS.
- W. Dzwolak, R. Ravindra and R. Winter, Phys. Chem. Chem. Phys., 2004, 6, 1938 RSC.
- L. Nielsen, S. Frokjaer, J. F. Carpenter and J. Brange, J. Pharm. Sci., 2001, 90, 29 CrossRef CAS.
- W. Dzwolak, R. Ravindra, J. Lendermann and R. Winter, Biochemistry, 2003, 42, 11347 CrossRef CAS.
- W. Dzwolak, V. Smirnovas, R. Jansen and R. Winter, Protein Sci., 2004, 13, 1927 CrossRef CAS.
- L. Nielsen, R. Khurana, A. Coats, S. Frokjaer, J. Brange, S. Vyas, V. N. Uversky and A. L. Fink, Biochemistry, 2001, 40, 6036 CrossRef CAS.
- S. N. Timasheff, Proc. Natl. Acad. Sci. USA, 2002, 99, 9721 CrossRef CAS.
- W. Dzwolak, R. Ravindra, C. Nicolini, R. Jansen and R. Winter, J. Am. Chem. Soc., 2004, 126, 3762 CrossRef CAS.
- P. Chien, J. S. Weissman and A. H. DePace, Annu. Rev. Biochem., 2004, 73, 617 CrossRef CAS.
- A. T. Petkova, R. D. Leapman, Z. H. Guo, W. M. Yau, M. P. Mattson and R. Tycko, Science, 2005, 307, 262 CrossRef CAS.
| This journal is © the Owner Societies 2005 |
