Protein immobilization on carbon nanotubes via a two-step process of diimide-activated amidation
Kuiyang
Jiang
*a,
Linda S.
Schadler
a,
Richard W.
Siegel
a,
Xinjie
Zhang
b,
Haifeng
Zhang
c and
Mauricio
Terrones
d
aDepartment of Materials Science and Engineering and Rensselaer Nanotechnology Center, Rensselaer Polytechnic Institute, Troy, NY 12180, USA. E-mail: kyjiang@yahoo.com
bPacific Northwest National Laboratory, Richland, WA 99352, USA
cDepartment of Physics, Washington State University, Richland, WA 99352, USA
dAdvanced Materials Department, IPICyT, Av. Venustiano Carranza 2425-A, Colonia Bellas Lomas, 78210, San Luis Potosi, SLP, Mexico
First published on 6th November 2003
Abstract
Ferritin and bovine serum albumin (BSA) proteins are chemically bonded to nitrogen-doped multi-walled carbon nanotubes (CNx MWNTs) through a two-step process of diimide-activated amidation. First, carboxylated CNx MWNTs were activated by N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDAC), forming a stable active ester in the presence of N-hydroxysuccinimide (NHS). Second, the active ester was reacted with the amine groups on the proteins of ferritin or BSA, forming an amide bond between the CNx MWNTs and proteins. This two-step process avoids the intermolecular conjugation of proteins, and guarantees the uniform attachment of proteins on carbon nanotubes. TEM and AFM measurements clearly confirmed the successful attachment. This approach provides a universal and efficient method to attach biomolecules to carbon nanotubes at ambient conditions.
Carbon nanotubes exhibit interesting electrical, structural and mechanical properties that make them highly promising nanoscale building blocks for the construction of novel functional materials. Many potential applications have been proposed, such as conductive and high-strength composites, field emission displays, fuel cells, sensors, and hydrogen storage media.1,2 In addition, biosensors for detecting abnormalities3–5 and bio-fuel cells6 for embedded devices are among the most exciting applications. In order to create the synergy between the biomolecules and nanotubes required to realize these applications, biomolecules, such as proteins and DNAs, must be connected to the carbon nanotubes. This connection can be non-covalent interaction or covalent bonding. There have been several reports on the immobilization of biomolecules on carbon nanotubes,7–13 and most of them use non-covalent interaction. The best stability, accessibility and selectivity, however, will be achieved through covalent bonding because of its capability to control the location of the biomolecule, improve stability, accessibility and selectivity and reduce leaching. In the present study, we report the covalent bonding of proteins to nitrogen-doped multiwalled carbon nanotubes (CNx MWNTs) via a two-step process of diimide-activated amidation between the carboxylic acid groups on CNx MWNTs and the amine groups on proteins.
To covalently bond molecules to the nanotubes first requires the formation of functional groups on the carbon nanotubes. The carboxylic acid group is often the best choice because it can undergo a variety of reactions and is easily formed on carbon nanotubes via oxidizing treatments, e.g., sonication in sulfuric and nitric acid, refluxing in nitric acid, ozonolysis, and air oxidization. The control of reactants and/or reaction conditions may control the locations and density of the carboxylic groups on the nanotubes, which can be used to control the locations and density of the attached biomolecules. One of the universal methods for connecting biomolecules to other materials is diimide-activated amidation, by direct coupling of carboxylic acid to proteins using N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDAC) or N,N′-dicyclohexylcarbodiimide (DCC) as a coupling agent. However, this process leads to undesirable side reactions of intermolecular conjugation of proteins, because most proteins are rich in both amine groups and carboxylic acid groups on their surface. This intermolecular connection can be avoided by using a two-step process: carboxylic acid groups are first converted to active esters via diimide-activation, and then the active esters are reacted with the amine groups on proteins without the presence of diimide. Thus, the process can guarantee homogenous attachment of proteins onto carbon nanotubes.
Fig. 1 shows the overview of our covalent attachment process. CNx MWNTs were acid-oxidized to form carboxylic acid groups on the surface of the carbon nanotubes. Then, the carboxylic acid groups were activated by EDAC, forming a highly reactive O-acylisourea active intermediate. The intermediate is unstable in aqueous solution, and does not have a sufficient lifetime for the two-step conjugation procedure. However, in the presence of N-hydroxysuccinimide (NHS), a more stable active ester (succinimidyl intermediate) can be formed. The active ester undergoes nucleophilic substitution reaction with the amine groups on proteins, resulting in the formation of an amide bond between the CNx MWNTs and proteins.
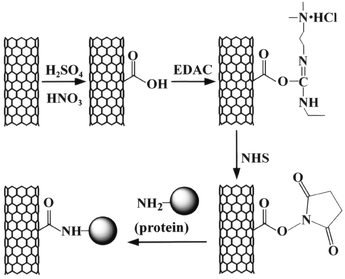 | ||
| Fig. 1 Schematic view of the attachment of proteins to carbon nanotubes via a two-step process of diimide-actived amidation. | ||
CNx MWNTs were synthesized by pyrolyzing ferrocene/melamine mixtures at 1050 °C in an Ar atmosphere.14 The as-produced material consists of carpet-like structures containing highly oriented nanotubes of uniform diameter (ca. 10–50 nm OD) and length (ca. 50–60 µm). The as-produced CNx MWNTs were suspended in a concentrated sulfuric acid–nitric acid mixture (3∶1 v/v) and sonicated in a sonication bath for 2 h. A nanotube mat was obtained after filtration using a 0.45 µm hydrophilized PTFE membrane and washed with deionized water until no acid was detected, followed by drying under vacuum. After the acid-treatment, the CNx MWNTs dispersed easily in water after sonication for a short period of time. TEM measurements indicated that the acid-treated nanotubes were well separated and shortened (ca. 5–10 µm). FT-IR studies revealed that this acid-treatment generated four types of functional groups on the CNx MWNTs: hydroxyl groups (3424 cm−1), carboxyl groups (1719 cm−1), carbonyl groups (1626 cm−1) and sulfate groups (1384 cm−1).15
The two-step attachment process is described in detail here. In the first step, 2.0 mg of carboxylated CNx MWNTs were suspended in 5.0 ml of deionized water by sonicating the mixture for a short period of time. Then, 1.0 ml of a 500 mM MES buffer solution (pH = 6.1) and 2.3 ml of a 50 mg ml−1 NHS aqueous solution were added to the above suspension and mixed. Under fast stirring, 1.2 ml fresh EDAC aqueous solution (10 mg ml−1) was added quickly, and the mixture was continually stirred at room temperature for 30 min. The suspension was then filtered through a 0.45 µm hydrophilized PTFE membrane and rinsed thoroughly with a 50 mM MES buffer solution (pH = 6.1) to remove excess EDAC, NHS and byproduct urea.
In the second step, the estered carbon nanotubes were re-dispersed in 9.0 ml of a 50 mM MES buffer solution (pH = 6.1) and 1.0 ml of a 10 mg ml−1 protein in MES buffer solution (pH = 6.1) was added. After shaking the mixture on a platform shaker at 150 rpm at room temperature for 1 h, the nanotube suspension was centrifuged and washed with 50 mM MES buffer solution (pH = 6.1) three times to remove unbound protein. The washed protein–nanotube conjugates were dispersed in 50 mM MES buffer solution (pH = 6.1) or deionized water for TEM or AFM measurements. Two kinds of proteins, ferritin from horse spleen and bovine serum albumin (BSA), were attached to CNx MWNTs in this study.
TEM and AFM studies confirmed the successful immobilization of both ferritin and BSA onto CNx MWNTs by the aforementioned two-step process. Fig. 2 shows a typical TEM micrograph of ferritin on CNx MWNTs. Ferritin molecules densely decorate the sidewalls of the carbon nanotubes, and the iron core of each ferritin (∼7 nm) can be clearly observed. The decoration is quite uniform and exists as a submonolayer. This indicates that the intermolecular aggregation of the proteins is negligible. The homogeneous positioning of ferritin on the surfaces of the carbon nanotubes is very similar to the attachment of Au particles onto carbon nanotubes that we previously reported.15 The location of the ferritins is representative of where the carboxylic acid groups were. Because the locations and density of carboxylic acid groups on the carbon nanotubes can be controlled by the oxidization and reaction conditions, we can control the locations and density of the attached biomolecules, which is extremely useful in some applications for high sensitivities.16
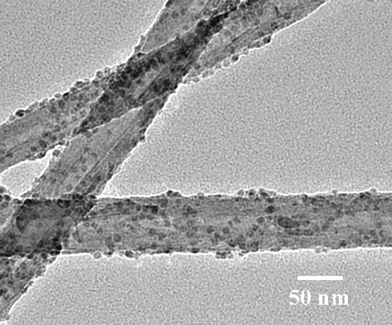 | ||
| Fig. 2 TEM image of ferritin-CNx MWNT conjugates. | ||
To confirm the protein immobilization mechanism outlined in Fig. 1, we carried out two control experiments without adding EDAC or NHS. The control experiment without adding EDAC showed that the carbon nanotubes were rather clean with little attachment of ferritin, and the control experiment without adding NHS showed that ferritin aggregates existed and that the carbon nanotubes were almost free of absorbed ferritin on their sidewalls.
Fig. 3 shows representative AFM images of the attached BSA on carbon nanotubes. In Fig. 3(a), a submonolayer of BSA proteins densely decorates the carbon nanotubes and no aggregation is obviously observed. The BSA molecules retain their original globular conformation, and their size estimated from the image is 30 nm. The size discrepancy with the hydrodynamic prolate ellipsoidal shape of BSA, 14 × 4 nm,17 is attributed to an AFM tip-broadening effect. In Fig. 3(b), the density of BSA molecules on the sidewall of the carbon nanotube is lower than that in Fig. 3(a). The reason for this is that carbon nanotubes with small diameter may contain fewer structural defects, so they will form fewer carboxylic acid groups through acid-treatment, since carboxylic acid groups are preferably derived from the structural defect sites.18,19 Also, carbon nanotubes with small diameter have higher surface curvature, which may not match well with the conformation of BSA molecules.
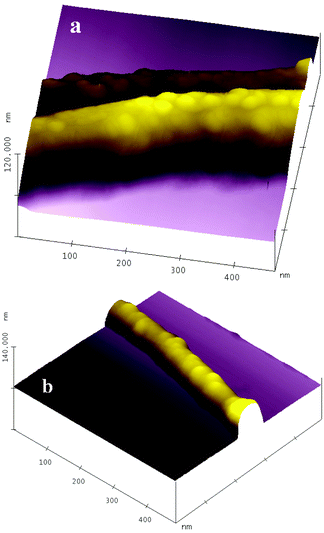 | ||
| Fig. 3 AFM images of BSA-CNx MWNT conjugates. | ||
In summary, we have demonstrated a useful, simple and universal method to attach biomolecules onto carbon nanotubes with covalent bonding. The two-step process was carried out at room temperature in buffer solutions and was accomplished in a short time, which will maximize the survival rate of biomolecules. This method can be easily applied to attach any other useful entities with amine groups, such as nucleic acids, polymers, dendrimers and even inorganic nanoparticles on carbon nanotubes. It is expected that this approach will provide novel nanoscale building blocks for the construction of biosensors, biocatalysts, bio-fuel cells, and other relevant structures.
Acknowledgements
The authors thank Dr Chris Aardahl and Dr Jungbae Kim for helpful discussions, and thank Dr Chongming Wang and Mrs Alice Dohnalkova for their help on TEM measurements. The work was performed at EMSL, a national scientific user facility sponsored by DOE’s Office of Biological and Environmental Research and located at PNNL, operated for DOE by Battelle. The work was supported in part by the U. S. Army Natick Soldier Systems Center under Contract No. DAAD 16-00-C-9237 and in part by the Nanoscale Science and Engineering Initiative of the National Science Foundation under NSF Award No. DMR-D117792.References
- R. H. Baughman, A. A. Zakhidov and W. A. de Heer, Science, 2002, 297, 787 CrossRef CAS.
- H. J. Dai, Acc. Chem. Res., 2002, 35, 1035 CrossRef CAS.
- C. V. Nguyen, L. Delzeit, A. M. Cassell, J. Li, J. Han and M. Meyyappan, Nano Lett., 2002, 2, 1079 CrossRef CAS.
- S. C. Tsang, Z. J. Guo, Y. K. Chen, M. L. H. Green, H. A. Hill, T. W. Hambley and P. J. Sadler, Angew. Chem., Int. Ed. Engl., 1997, 36, 2198 CAS.
- K. A. Willams, P. T. M. Veenhuizen, B. G. de la Torre, R. Eritja and C. Dekker, Nature, 2002, 420, 761 CrossRef.
- I. Willner, Science, 2002, 298, 2407 CrossRef CAS.
- S. C. Tsang, J. J. Davis, M. L. H. Green, H. A. O. Hill, Y. C. Leung and P. J. Sadler, Chem. Commun., 1995, 1803 RSC.
- F. Balavoine, P. Schultz, C. Richard, V. Mallouh, T. W. Ebbesen and C. Mioskowski, Angew. Chem., Int. Ed., 1999, 38, 1912 CrossRef CAS.
- R. J. Chen, Y. Zhang, D. Wang and H. Dai, J. Am. Chem. Soc., 2001, 123, 3838 CrossRef CAS.
- W. Huang, S. Taylor, K. Fu, Y. Lin, D. Zhang, T. W. Hanks, A. M. Rao and Y. P. Sun, Nano Lett., 2002, 2, 311 CrossRef CAS.
- B. R. Azamian, J. J. Davis, K. S. Coleman, C. B. Bagshaw and M. L. H. Green, J. Am. Chem. Soc., 2002, 124, 12664 CrossRef CAS.
- K. Besteman, J. O. Lee, F.G. M. Wiertz, H. A. Heering and C. Dekker, Nano Lett., 2003, 3, 727 CrossRef CAS.
- B. F. Erlanger, B.-X. Chen, M. Zhu and L. Brus, Nano Lett., 2001, 1, 465 CrossRef CAS.
- M. Terrones, H. Terrones, N. Grobert, W. K. Hsu, Y. Q. Zhu, J. P. Hare, H. W. Kroto, D. R. M. Walton, P. Kohler-Redlich, M. Rühle, J. P. Zhang and A. K. Cheetham, Appl. Phys. Lett., 1999, 75, 3932 CrossRef CAS.
- K. Jiang, A. Eitan, L. S. Schadler, P. M. Ajayan, R. W. Siegel, N. Grobert, M. Mayne, M. Reyes-Reyes, H. Terrones and M. Terrones, Nano Lett., 2003, 3, 275 CrossRef CAS.
- J. Li, H. T. Ng, A. Cassell, W. Fan, H. Chen, Q. Ye, J. Koehne, J. Han and M. Meyyappan, Nano Lett., 2003, 3, 597 CrossRef CAS.
- T. Peters, Adv. Protein Chem., 1985, 37, 161 CAS.
- M. Monthioux, B. W. Smith, B. Burteaux, A. Claye, J. E. Fischer and D. E. Luzzi, Carbon, 2001, 39, 1251 CrossRef CAS.
- M. A. Hamon, H. Hu, P. Bhowmik, S. Niyogi, B. Zhao, M. E. Itkis and R. C. Haddan, Chem. Phys. Lett., 2001, 347, 8 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2004 |
