-
PDF
- Split View
-
Views
-
Cite
Cite
Nataliya Kitsera, Julia Allgayer, Edris Parsa, Nadine Geier, Martin Rossa, Thomas Carell, Andriy Khobta, Functional impacts of 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine at a single hemi-modified CpG dinucleotide in a gene promoter, Nucleic Acids Research, Volume 45, Issue 19, 2 November 2017, Pages 11033–11042, https://doi.org/10.1093/nar/gkx718
Close - Share Icon Share
Abstract
Enzymatic oxidation of 5-methylcytosine (5-mC) in the CpG dinucleotides to 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC) and 5-carboxycytosine (5-caC) has central role in the process of active DNA demethylation and epigenetic reprogramming in mammals. However, it is not known whether the 5-mC oxidation products have autonomous epigenetic or regulatory functions in the genome. We used an artificial upstream promoter constituted of one cAMP response element (CRE) to measure the impact of 5-mC in a hemi-methylated CpG on the promoter activity and further explored the consequences of 5-hmC, 5-fC, and 5-caC in the same system. All modifications induced mild impairment of the CREB transcription factor binding to the consensus 5′-TGACGTCA-3′ CRE sequence. The decrease of the gene expression by 5-mC or 5-hmC was proportional to the impairment of CREB binding and had a steady character over at least 48 h. In contrast, promoters containing single 5-fC or 5-caC underwent further progressive loss of activity, up to an almost complete repression. This decline was dependent on the thymine-DNA glycosylase (TDG). The results thus indicate that 5-fC and 5-caC can provide a signal for perpetuation and enhancement of the repressed transcriptional state by a mechanism that requires base excision repair.
INTRODUCTION
DNA methylation at cytosine residues plays essential roles in transcriptional regulation and genome maintenance in vertebrates (1–3). Most abundant and best studied context in which 5-methylcytosine is present in the genomes is symmetric methylation of CpG dinucleotides. CpG methylation can be copied to daughter DNA strands during replication and thus constitutes an epigenetic mark propagated through cell division (4,5). CpG methylation in the promoter regions regulates transcriptional activity of genes by at least two mechanisms. One is direct modulation of binding of the methylation-sensitive transcription factors to the regulatory motifs containing methylated CGs (6–8). Another is reinforcement and long-term propagation of chromatin-mediated gene silencing by recruitment of the methyl-CpG-binding domain (MBD) proteins and, in turn, histone deacetylases (1,2). Both these modes of action appear to be important for silencing and transcriptional repression of genes whose promoters contain long-range arrays rich in CpG dinucleotides, known as CpG islands (CGIs). In the non-CGI promoters, functions of CpG methylation are even more complex and the outcomes for transcriptional activity may be very variable for genes controlled by different transcription factors and in different cell lineages (9–11). DNA demethylation mechanisms add another complexity level to the regulation of gene transcription by erasing 5-methylcytosine or converting it to other types of base modifications whose regulatory functions still have to be explored (12–15). Thus, currently the best characterized DNA demethylation pathway in vertebrates mediated by the ten eleven translocation (TET) family oxygenases (16–21) generates 5-hmC, 5-fC and 5-caC, which may have independent functions in the regulation of gene expression (13,22). However, understanding of functional consequences of cytosine demethylation products at specific regulatory elements is hindered by the extremely low levels of these modifications in the genome (17,19,23) and by the dynamic character of 5-fC and 5-caC which are efficiently removed by DNA repair mechanisms (15,18,24).
Transitions between the methylated and unmethylated states of CpG dinucleotides require hemi-modified states in which 5-mC or its oxidation products are present in only one DNA strand (25). Functional impact of such modifications on the regulation of gene expression is unknown because deliberate hemi-modification of chosen cytosine residues in DNA cannot be generated enzymatically. To overcome this hurdle, we have previously proposed an efficient procedure for a site-specific incorporation of synthetic oligonucleotides containing various base modifications into plasmid-borne reporter genes with the help of sequence-specific nicking endonucleases (26). With these tools, we have already obtained important insights into the mechanisms of excision repair of several types of structurally defined DNA lesions in functional reporter genes (27–29). Here, we applied an analogous approach to measure the functional impacts of 5-mC, 5-hmC, 5-fC and 5-caC on the regulation of gene transcription. Our strategy was to generate expression constructs containing each of these cytosine modifications in a CpG dinucleotide within an artificial promoter composed of a cyclic adenosine monophosphate (cAMP) response element (CRE) upstream from a TATA box-containing RNA polymerase II basal promoter. We chose CRE because it contains a CpG dinucleotide in the consensus sequence and because it was known that binding of the specific transcription factors (e.g., CREB and C/EBP alpha) and the induced transcription can be either negatively or positively regulated by the symmetric CpG methylation (8,30). Moreover the presence of 5-mC and 5-hmC in just one DNA strand significantly reduce, whereas the C>U substitution in either one or both DNA strands increase the affinity of CREB binding (31,32). The aim of present work was to compare the functional significance of CpG hemi-methylation and hemi-hydroxymethylation for the CRE-driven gene expression and to investigate the impacts of the DNA demethylation intermediates 5-fC and 5-caC on the promoter activity.
MATERIALS AND METHODS
Cell lines and reporter vectors
Isogenic HeLa-derived cell lines in which TDG, SMUG1 or UNG1/2 DNA glycosylases were knocked down by stable expression of the specific shRNA and the corresponding control HeLa/pEpS cell line were generated and characterised by western blotting and the specific DNA glycosylase activity testing previously (29). The sequence cloned into the pENTR/pSuper+ vector (Addgene, Cambridge, MA, USA) for expression of shRNA which efficiently targets human TDG was 5′ GATCCGGGAACGAAATATGGACGTTCAACTCGAGTTGAACGTCCATATTTCGTTCTTTTTGGAAA. The parental vector pENTR/pSuper+ was used as a scrambled transcript control. Vectors pCRE-uno with a unique full-length 5′-TGACGTCA-3′ consensus CRE sequence upstream from the RNA polymerase II transcription initiation region of the reporter EGFP gene and pCMV1111 with four CRE sequences were described previously (33). The vectors contain tandem sites for the Nb.BsrDI nicking endonuclease spaced with the 18-nucleotide interval around the 5′-TGACGTCA-3′ consensus CRE sequence (Figure 1A). The derived vectors pCRE-uno-C (with inverse orientation of the Nb.BsrDI sites but otherwise identical to pCRE-uno) and pCRE-zero (in which CRE was mutated to produce an inert 5′-ACTGACTG-3′ sequence) were generated by PCR with the overlapping primers using PfuTurbo® polymerase (Agilent Technologies, Waldbronn, Germany). The template DNA was digested with DpnI (Thermo Fischer Scientific, Darmstadt, Germany) followed by direct repair in the ultracompetent Escherichia coli scs-8 (Agilent Technologies).
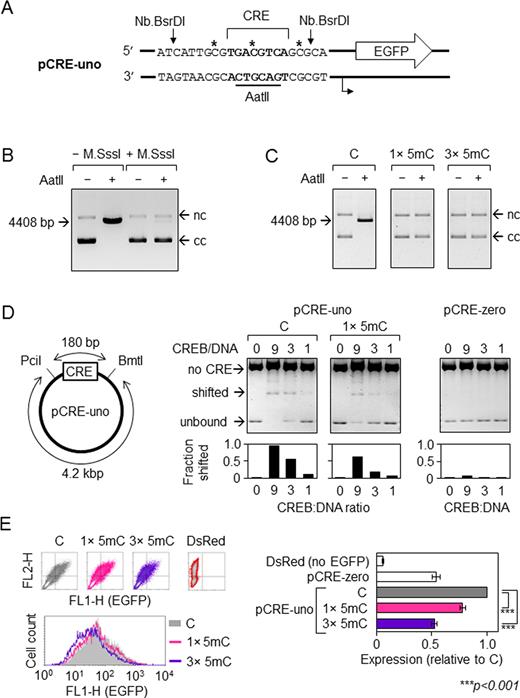
Impact of CpG hemi-methylation in the minimal CRE promoter on the CREB binding and the reporter EGFP gene expression. (A) Artificial CRE-uno promoter: transcription start (broken arrow), CRE sequence (bold), AatII site (underlined), Nb.BsrDI nicking positions (vertical arrows), and positions of 5-mC/5-hmC/5-fC/5-caC in the incorporated synthetic oligonucleotides (asterisks). (B) Inhibition of the AatII restriction endonuclease by the M.SssI methylation of the central CG-dinucleotide in CRE. Agarose gel electrophoresis of pCRE-uno incubated with AatII. Arrows indicate migration positions of the linearized vector (4408 bp) and of the covalently closed (cc) and nicked (nc) forms of circular vector DNA. (C) Verification of the incorporation of synthetic oligonucleotides containing one or three 5-mC (1 × 5mC, 3 × 5mC) by inhibition of the AatII cleavage. (D) Scheme of the PciI/BmtI digest of the pCRE-uno vector and electrophoretic mobility shift assay (EMSA) of the PciI/BmtI fragments under the indicated CREB:DNA molar ratios. The shorter (180 bp) fragment contains CRE with no or one 5-mC in the central CG-dinucleotide. The right panel shows control CRE-less vector (pCRE-zero) incubated with CREB in parallel. (E) EGFP expression in HeLa cells transfected with pCRE-uno containing none, one (in the central CG-dinucleotide of CRE) or three 5mC. Representative FACS data and relative EGFP expression values (mean ± SD, n = 7). Expression in the absence of an EGFP-coding vector (DsRed) denotes the lower detection limit. CRE-less vector (pCRE-zero) was included in two experiments as a reference for the basal expression level. P-values calculated by the Student's t-test.
Synthetic oligonucleotides
DNA CE-phosphoramidites Bz-dA, Bz-dC, iBu-dG, T and Bz-mC were obtained from Glen Research (Sterling, VA) or Link Technologies (Bellshill, Scotland). 5-HmC, 5-fC and 5-caC phosphoramidites were synthesized and solid-phase synthesis of the 18-mer 5′-CATTGCGTGACGTCAGCG deoxyribo-oligonucleotides was performed using the standard protocols described previously (22). Underlined cytosines show positions in which 5-mC/5-hmC/5-fC/5-caC were incorporated, as specified in the text. Synthesized DNA chains were HPLC-purified with VP 250/10 Nucleosil 100-7 C 18 columns (Macherey-Nagel, Düren, Germany) and the quality determined by MALDI-MS (22). 2′-(R)-fluorinated derivatives of 5-fC and 5-caC were synthetised and handled as described previously (34). The sequence for incorporation of the 2′-fluorinated nucleotides was 5′-CATTGCGTGACGTCAGCG. Oligonucleotides 5′-CATTGCGTGA[THF/S-THF]GTCAGCG containing the abasic site analog tetrahydrofuran with either phosphodiester (THF) or phosphorothioate (S-THF) 5′-linkage as well as 5′-CATTGCGTGAC[8-oxoG]TCAGCG containing 8-oxo-7,8-dihydroguanine were purchased from BioSpring GmbH (Frankfurt am Main, Germany).
Generation of the reporter constructs with 5-mC/5-hmC/5-fC/5-caC specifically positioned in the CRE sequence
Site-specific double nicks on both sides of the CRE sequence in the vectors pCRE-uno (‘top’ strand), pCRE-uno-C (‘bottom’ strand) or pCMV1111 were generated by the Nb.BsrDI endonuclease (NEB GmbH, Frankfurt am Main, Germany). Synthetic 18-mer oligonucleotides containing 5-mC/5-hmC/5-fC/5-caC, the 2′-(R)-fluorinated derivatives of 5-fC/5-caC or THF/S-THF in the specified positions were used to displace the excised native DNA strand fragment and seamlessly ligated into vector DNA by the strand exchange protocol described previously (26). The respective control oligonucleotide without modifications was always ligated in parallel for every independent vector preparation. The presence of 5-mC/5-hmC/5-fC/5-caC in the covalently closed vector DNA was verified by the inhibition of cleavage by the AatII restriction endonuclease (NEB).
Electrophoretic mobility shift assays
Vectors pCRE-uno and pCRE-zero were digested with PciI and BmtI (both NEB) to produce the CRE-less 4228 bp fragment and the 180 bp fragment with one (pCRE-uno) or no CRE (pCRE-zero) and cleaned up. Binding reactions contained 10 nM DNA (400 ng in 15 μl reaction volume) and proportional variable amounts of the purified CREB protein (BioCat, Heidelberg, Germany) in the binding buffer composed of 20 mM Tris-HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol and 5% glycerol (all reagents from Sigma-Aldrich, Seelze, Germany) supplemented with 1 mg/ml bovine serum albumin (NEB). After 30-min incubation at 37°C reactions were chilled on ice and fragments separated by agarose gel electrophoresis in the 0.5 × TBE buffer (Bio-Rad Laboratories, Dreieich, Germany) under cooling conditions, as described previously (35). Band intensities were quantified following the ethidium bromide staining with the help of GelDocTM XR+ molecular imager and the Image LabTM software (Bio-Rad).
Transfections and gene expression analyses
Exponentially growing in six-well plates cells were transfected with the help of Effectene (QIAGEN, Hilden, Germany) with the EGFP reporter constructs (with or without the specified modifications) in combination with the tracer pDsRed-Monomer-N1 vector (Clontech, Saint-Germain-en-Laye, France), 400 ng each vector. The method for quantitative determination of EGFP expression in transiently transfected cells by flow cytometry was validated by titration of the EGFP-encoding vector and described in detail previously (36). Briefly, formaldehyde-fixed cells were equilibrated in phosphate buffered saline (PBS) and analysed using FACSCalibur™ and the CellQuest™ Pro software (Beckton Dickinson GmbH, Heidelberg, Germany). FSC/SSC gating was routinely used to exclude fragmented and aggregated cells and DsRed signal (FL2-H) was applied as additional gating marker to select for the effectively transfected cells. After the exclusion of untransfected cells, EGFP fluorescence (FL1-H) distribution plots were generated and average EGFP expression per cell determined as the median of the distribution. To assess variability between independent experiments, expression levels of constructs with cytosine modifications were calculated in each experiment relative to the expression of the control construct (without modifications) as a ratio of the respective FL1-H values.
RESULTS
Generation of expression constructs containing the C5 atom modifications of defined cytosines
We have previously generated the EGFP-encoding pCRE-uno reporter vector in which the basal RNA polymerase II promoter is coupled with a single CRE as a minimal upstream regulatory element. The CRE-motif is flanked by strand-specific nicking sites for the endonuclease Nb.BsrDI (Figure 1A), which are neutral for the regulation of gene expression, whereas CRE itself measurably activates transcription of the reporter EGFP gene (33). We sought to use these properties for the investigation of consequences of defined cytosine modifications at a single CpG dinucleotide for the regulation of the CRE-driven gene expression. For this purpose, we have introduced 5-mC (or 5-hmC/5-fC/5-caC) into the vector DNA sequence by substitution of the Nb.BsrDI-excised native DNA strand fragment for the synthetic strands containing the respective modifications at the central CG-dinucleotide of the CRE sequence.
As a proof of principle, we first generated expression constructs containing one or three 5-mC residues in the substituted DNA strand (Figure 1A). The pCRE-uno CRE sequence accommodates a unique recognition sequence for the AatII restriction endonuclease, whose activity is inhibited by the CpG methylation (Figure 1B), which enables easy verification of the incorporation of 5-mC with the synthetic oligonucleotide (Figure 1C). Subsequently, we used the same approach for the incorporation of oligonucleotides containing 5-hmC, 5-fC or 5-caC and demonstrated that all the modifications were efficiently integrated into vector DNA (Supplementary Figure S1).
CREB binding and gene expression in the presence of the CpG hemi-methylation
We have analysed by the electrophoretic mobility shift assay the influence of single 5-mC in the central CpG dinucleotide of the CRE consensus sequence on binding of the transcription factor CREB (Figure 1D). Comparison with the nearly identical vector pCRE-zero in which the CRE site has been deleted showed that CREB binds specifically to the restriction fragment containing the CRE-motif and that single 5-mC in the central CpG dinucleotide significantly reduces the band shifting. The equivalent levels of band shifting in the presence of 5-mC, were achieved by increasing the CREB:DNA ratio by the factor of ∼2.5 (Figure 1D and not shown data), suggesting that single methyl group causes a 2- to 3-fold reduction of CREB affinity to its binding site.
To check whether the impaired CREB binding has a functional significance for the gene transcription in cells, we next measured the impact of CpG hemi-methylation on the EGFP expression levels in transfected human cells. Analyses in HeLa cells showed that single 5-mC incorporated into the central CpG nucleotide decreased the expression by approximately one quarter. Hemi-methylation of all three CpG dinucleotides within the exchanged DNA fragment resulted in the halved expression level with respect to the expression of unmethylated control templates (Figure 1E). This quantitatively corresponded to the expression level of the CRE-less vector, suggesting that hemi-methylation at these three positions reduces gene transcription to the basal levels and, hence, nullifies the activatory effect of CREB. Accordingly, we derive that hemi-methylation of the central CpG alone caused an ∼2-fold reduction of the CREB-dependent fraction of transcription. For comparison, total methylation of the pCRE-uno vector by M.SssI almost completely obliterated the EGFP expression, thus indicating a very efficient repression of both induced and basal transcription (Supplementary Figure S2). Together, the results indicate that even a single methyl group—if present at the critical CpG dinucleotide—can measurably reduce the level of gene transcription in cells by direct reduction of the affinity of the transcription factor binding.
The impact of 5-hmC on the gene expression: comparison with 5-mC
We have next generated expression constructs containing synthetic 5-hmC at the same CpG dinucleotides as described above for 5-mC and determined the expression levels in Hela cells under the same conditions. As 5-mC, 5-hmC caused a significant reduction in the EGFP expression levels if present in the central CpG dinucleotide of the CRE sequence and even stronger reduction when all three CpG dinucleotides of the incorporated synthetic DNA were modified by the hydroxymethyl group (Figure 2A). Quantitatively, these effects were similar to the magnitude of the inhibitory effects of 5-mC at the same positions. CREB binding to the modified CRE was also impaired by single 5-hmC to at least the same extent as by 5-mC (Figure 2B), which suggests that the observed decrease in the gene transcription levels in cells was caused primarily by the impaired binding of the transcriptional activator. It has to be noted, however, that the presence of additional methyl or hydroxymethyl groups at the CpG sites outside from the minimal consensus CRE sequence did not cause further prominent decrease of CREB binding in the band-shifting assays (Supplementary Figures S3 and S4), thus indicating that the binding mode and the impact of 5-hmC on the gene expression in cells may be additionally influenced by other factors.
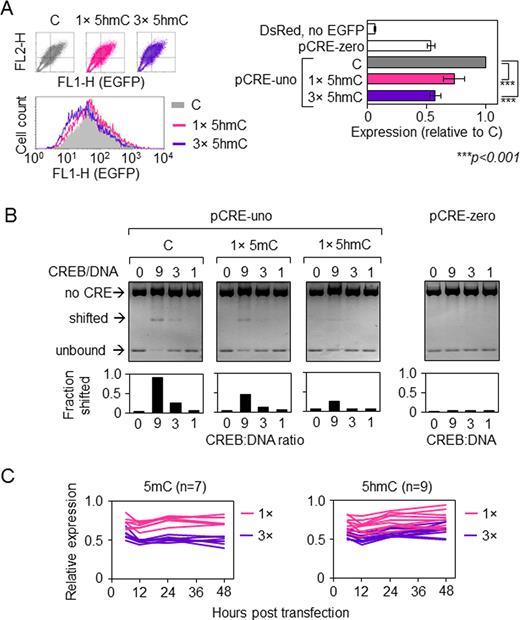
Impacts of one or three 5-hmC in the minimal CRE promoter on the EGFP gene expression and comparison with the respective effects of 5-mC. (A) EGFP expression in HeLa cells transfected with pCRE-uno containing none, one or three 5-hmC. Representative fluorescence scatter plots and the correspondent overlaid fluorescence distribution plots of HeLa cells 24 h after transfection (on the left) as well as mean EGFP expression values determined relative to the construct containing unmodified oligonucleotide (‘C’). Mean of nine independent experiments ± SD; P-values calculated by the Student's t-test. (B) CREB binding to CRE containing one 5-mC or 5-hmC in the central CG-dinucleotide detected by EMSA of the pCRE-uno PciI/BmtI fragment. (C) EGFP expression in HeLa cells transfected with pCRE-uno containing either one (1×) or three (3×) of the indicated CpG modifications in the promoter fragment (relative to pCRE-uno without CpG modifications). Lines represent independent transfections.
Because 5-hmC has been proposed to play a functional role in DNA demethylation (by either TDG-dependent (16,17,21) or -independent (37,38) pathways, we wondered whether the magnitudes of the repressory effects of both CpG modifications (5-mC and 5-hmC) would vary in the course of time. However, the results showed that the effects of both modifications persisted for at least 48 hours post transfection. During the whole time course, constructs with 5-mC and 5-hmC showed on average equal extents of transcriptional inhibition (Figure 2C), suggesting that 5-hmC has not been removed from CRE. The values for 5-hmC had a somewhat higher interexperimental noise; however, there was no clear tendency to recovery of the gene expression levels over the time course. The results thus indicate that, at least in the cell model used, hemi-methylated and hemi-hydroxymethylated CpG dinucleotides have overall very similar negative impacts on CREB binding and the CRE-activated transcription.
The effects of 5-mC and 5-hmC are manifested in both DNA strands
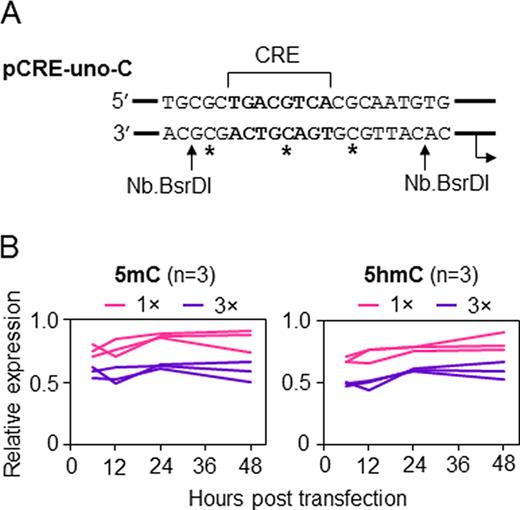
It is not known, whether biological consequences of hemi-methylation (or hemi-hydroxymethylation) depend on the DNA strand which is methylated. Because only one strand of the pCRE-uno plasmid (Figure 1A, top strand) can be substituted for a synthetic strand, we have generated an analogous pCRE-uno-C plasmid with flipped BsrDI sites, in which the complementary DNA strand can be replaced (Figure 3A, bottom strand). With CRE sequence as a reference point, we introduced 5-mC and 5-hmC at exactly the same CpG dinucleotides as previously in the top strand and performed time course expression analyses in HeLa cells (Figure 3B). Analogously to the previous experiments (Figure 2C), the results showed that either 5-mC or 5-hmC in the central CpG of the CRE sequence resulted in losses of approximately one fourth of the total gene expression level. Additional modifications at the neighbouring CpG dinucleotides caused ulterior decrease of the gene expression resulting in the loss of approximately half of the total transcriptional activity, which corresponds to residual activity equivalent to the basal transcription level of the CRE-less vector described above. Once again, the effects of both modifications persisted for at least 48 hours. Overall, we conclude that the effects of 5-mC and 5-hmC at the same hemi-modified CpG dyads on the transcriptional activation/repression of the CRE-regulated promoter are qualitatively and quantitatively very similar, regardless of the DNA strand affected by the modification.
Impacts of 5-mC and 5-hmC in the ‘bottom’ DNA strand of the minimal CRE promoter on the EGFP gene expression. (A) Scheme of the promoter fragment of the pCRE-uno-C vector used for modification of CG-dinucleotides in the bottom strand (asterisks). (B) EGFP expression in HeLa cells transfected with pCRE-uno containing either one (1×) or three (3×) of the indicated modifications in the promoter fragment (relative to pCRE-uno without modifications). Lines represent independent transfections.
Potent and largely indirect repression by 5-fC and 5-caC
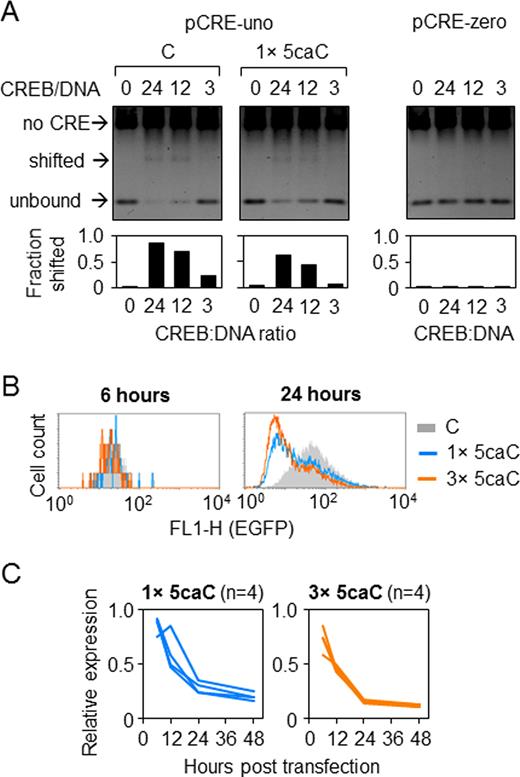
The TET-induced mechanism of enzymatic DNA demethylation described previously (16–21) foresees transient generation of 5-fC and perhaps 5-caC. Although active DNA demethylation is regarded as a potential mechanism for re-expression of genes originally silenced by DNA methylation, functional impact of these oxidation products on gene expression has not been sufficiently characterized to date. We have used the experimental setup described above to characterize the impacts of these cytosine modifications on the CRE-driven gene transcription. Because of the variable band-shifting activity between different batches of the recombinant CREB protein, we had to adjust the protein amounts in these experiments with respect to the conditions used in Figures 1D and 2B in order to achieve measurable band shifting with unmodified (‘C’) DNA (Figures 4A and 5A). We found that both 5-fC and 5-caC to some extent interfere with CREB-binding in vitro, however, do not prevent the binding completely, i.e. CREB binding properties to its target sequence were altered by these modifications to similar extents as earlier observed for hemi-methylated and hemi-hydroxymethylated CpG.
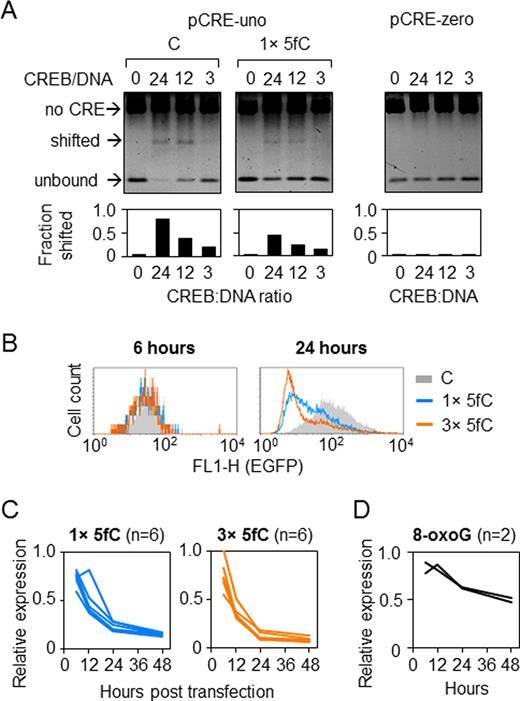
Effects of 5-fC in the minimal CRE promoter. (A) CREB binding to CRE containing one 5-fC in the central CG-dinucleotide detected by EMSA. (B) Representative fluorescence distribution plots of HeLa cells 24 h after transfection with constructs containing 5-fC in one (1×) or three (3×) CG-dinucleotides (overlaid with the reference ‘C’ construct obtained with unmodified synthetic oligonucleotide). (C) Time-course of the EGFP expression (relative to ‘C’) in HeLa cells transfected with constructs containing 1× or 3 × 5-fC. Lines show independent transfections. Mean values for each time point are highly significantly different from 1 (P < 0.001, heteroscedastic Student's t-test). (D) Time-course of the EGFP expression in HeLa cells transfected with constructs containing single 8-oxoG in the CRE (two independent experiments).
Effects of 5-caC in the minimal CRE promoter. (A) CREB binding to CRE containing one 5-caC in the central CG-dinucleotide detected by EMSA. (B) Representative fluorescence distribution plots of HeLa cells 24 h after transfection with constructs containing 5-caC in one (1×) or three (3×) CG-dinucleotides overlaid with the reference ‘C’ construct. (C) Time-course of the EGFP expression (relative to ‘C’) in HeLa cells transfected with constructs containing 1× or 3 × 5-caC. Lines show independent transfections. Mean values for each time point are highly significantly different from 1 (P < 0.001, heteroscedastic Student's t-test).
Expression analyses of the constructs containing one or three 5-fC in and around the CRE sequence showed that this modification leads to decreased reporter gene expression in the transfected HeLa cells (Figure 4B and C). The impairment of the EGFP expression was initially very mild (as judged from the fluorescence measured at the earliest time point of 6 h); however, the magnitude of the negative impact of 5-fC on the gene expression greatly increased by the 24 h time point after transfection (Figure 4B). At 24 h post-transfections, the degrees of the inhibition of gene expression by 5-fC and 5-caC clearly exceeded the respective effects of 5-mC and 5-hmC reported above (Figures 1–3). In-depth monitoring of the expression of constructs containing one or three hemi-modified CpG sites over a time span of 6–48 h post-transfection revealed a very strong and steady decline of the EGFP expression with no signs of recovery over the whole observation period, suggesting that 5-fC induced persistent repression of the gene transcription (Figure 4C). Thereby, the gene repression could not be accounted solely to the inhibition of CREB binding, as concluded from both strength and kinetics of the observed effects.
It is interesting to note that a similar gradual transcriptional repression was previously reported for reporter constructs containing various nucleobase modifications and has been identified as a hallmark of DNA lesions undergoing base excision repair (BER) (27,29,33). The role of BER in the gene repression has been inferred based on the requirements of the lesion-specific DNA glycosylases and of the strand-cleaved reaction intermediate generated by the apurinic/apyrimidinic site endonuclease APE1 (33). To test whether BER of a modification situated in the CRE sequence would affect the gene expression in a comparable way, we have placed single 8-oxo-7,8-dihydroguanine (8-oxoG) as a representative BER substrate into the CRE sequence. The results indeed showed a time-dependent decline of the gene expression (Figure 4D). Thus, 8-oxoG and 5-fC in the CRE sequence induced qualitatively similar transcriptional responses; however, the negative impact of 5-fC on the gene expression was much more powerful.
Further, we have analysed the CREB binding (Figure 5A) as well as the EGFP expression (Figure 5B and C) with 5-caC incorporated into the pCRE-uno reporter vector. The results showed that responses to 5-caC essentially recapitulated the response to 5-fC, i.e. mild inhibition of CREB binding and time-dependent progressive repression of transcription in transfected HeLa cells. Together, the results indicate that transcriptional responses to 5-fC and 5-caC largely resemble the response to DNA lesions, as exemplified by 8-oxoG. The observed transcriptional repression of the reporter constructs containing 5-fC or 5-caC further suggests that also oxidation of 5-mC/5-hmC to 5-fC/5-caC in the process of TET-induced DNA demethylation may not necessarily lead to transcriptional re-activation.
TDG mediates BER-dependent transcriptional repression by 5-fC and 5-caC
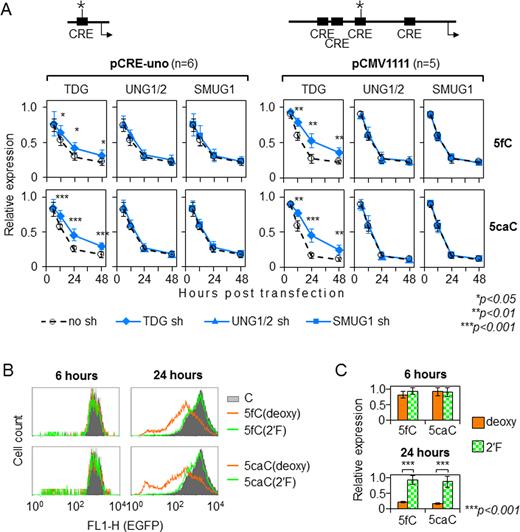
Because BER intermediate has been previously implicated in transcriptional repression or silencing induced by various DNA lesions (27,29,33,36), we questioned whether the progressive inhibition of expression of the reporter vectors containing 5-fC or 5-caC is similarly mediated by BER. Both 5-fC and 5-caC are efficiently removed from DNA in vivo and in vitro by TDG (16,18); therefore we performed gene expression analyses of vectors containing these modifications in cells with knocked down TDG expression (Supplementary Figure S5). The negative effects of 5-caC in pCRE-uno on the reporter gene expression were greatly reduced in the cell line stably expressing the TDG-specific short hairpin (sh) RNA but not in the isogenic cell lines with knocked down UNG1/2 or SMUG1 DNA glycosylases and not in the cell line stably transfected with non-specific shRNA (Figure 6A, left panel). The results thus indicate that excision of 5-caC by TDG is a prerequisite of ulterior transcriptional silencing of the CRE-controlled expression constructs. The magnitude of the inhibitory effects of 5-fC on the gene expression was also significantly moderated by the TDG knockdown, however to a lesser extent than it was observed for 5-caC.
To verify whether the effects of TDG knockdown were specific to 5-fC/5-caC, we further generated expression constructs carrying AP site (THF) in the same position in CRE (Supplementary Figure S6). As expected, the expression analyses showed a strong decrease of EGFP expression induced by THF but not by the APE1-resistant AP site (S-THF). Importantly, the magnitude of the effect of THF was not at all alleviated by the TDG knockdown. We thus conclude that BER reactions downstream from the base excisions step were not affected.
Impact of BER on the expression of reporter constructs containing single 5-fC/5-caC in the CRE sequence. (A) Effects of 5-fC and 5-caC on EGFP expression in HeLa-derived cell lines with stable shRNA knockdown of the UNG1/2, TDG or SMUG1 DNA glycosylases as compared to the control isogenic cell line (‘no sh’). Expression of the constructs containing 5-fC or 5-caC in the pCRE-uno (left panel) and pCMV1111 (right panel) vectors at each time point is presented relative to the respective reference constructs containing cytosine (mean ± SD; P-values calculated by the Student's t-test). (B, C) EGFP expression in HeLa cells at the specified times after transfection with pCMV1111 containing single 5-fC or 5-caC as deoxyribonuceotides (deoxy) or the respective 2′-(R)-fluorinated derivatives (2′F). Representative FACS data (B) and relative EGFP expression values (mean ±SD, n = 5, P-values calculated by the Student's t-test) (C).
Because TDG knockdown had quantitatively only modest impact on the expression of constructs containing 5-caC and especially 5-fC, we sought to increase the dynamic range for measurements of the EGFP expression. Therefore, we performed analogous experiments in the pCMV1111 vector which contains a strong promoter with multiple regulatory elements including four consensus CRE sequences, one of which was chosen for targeted incorporation of synthetic C/5-fC/5-caC (Supplementary Figure S1B). Remarkably, also in the context of the strong promoter, both 5-caC and 5-fC induced dynamic decrease of the gene expression and in both cases the effects were significantly moderated by the TDG knockdown (Figure 6A, right panel). Also in this promoter the effect of TDG knockdown was somewhat smaller for 5-fC than 5-caC, but the difference was far less pronounced than in pCRE-uno.
The results obtained in the TDG knockdown cells underpinned the role of BER of 5-fC and 5-caC in the transcriptional repression by these cytosine modifications (Figure 6A); however, since the recovery of gene expression was not complete, they did not exclude a potential contribution of other mechanisms. To eliminate the base excision activity completely, we have applied a chemical approach. It was previously reported that sugar 2′-fluorination of the 5-fC and 5-caC nucleotides prevents the TDG excision activity completely while having only minimal impact on the physical and functional properties of the DNA helix (34). Therefore, we next generated the pCMV1111 expression constructs carrying single 5-fC/5-caC deoxyribonucleotides or their 2′-fluorinated analogs in the specified position in CRE (Supplementary Figure S7). The expression analyses in HeLa cells explicitly showed that 2′-fluorination of deoxyribose fully reversed the gene repression by 5-fC and 5-caC (Figure 6B and C). This effect was exactly reproduced in the pCRE-uno vector (Supplementary Figure S8). The results thus show that regulatory promoter elements retain their activatory functions and that 5-fC and 5-caC do not lead to transcriptional repression when the base excision is inhibited, e.g. by the deoxyribose 2′-fluorination.
DISCUSSION
There are at least two mechanisms for regulation of gene transcription by 5-mC in DNA. It was previously suggested that selective methylation of CpG dinucleotides present in upstream promoter elements can interfere with recruitment of the specific transcription factors (6–8). On the other hand, CpG methylation can recruit general co-repressors via the MBD binding (1,2). Using a minimal CRE-driven promoter with an artificially introduced hemi-methylated CpGs in defined positions, we show here that a single methyl group in the central CpG of the CRE sequence causes a 2- to 3-fold reduction of the specific CREB binding affinity and an equivalent decrease in the CRE-dependent gene expression in human cells (Figure 1). This proportionality suggests that impaired CREB binding has direct functional relationship with the gene expression. Nevertheless, hemi-methylation of the neighboured CpGs clearly added to the decreased gene expression in cells (Figures 2 and 3), even though CREB binding was not further impaired by the modifications outside of the CRE consensus sequence (Figures 2 and 3, Supplementary Figures S3 and S4), which indicates that impaired CREB binding is not the only mechanism for the declined gene expression if more than one 5-mC is present. Indeed, massive methylation of the reporter vector by M.SssI resulted in a complete transcriptional silencing of the reporter constructs (Supplementary Figure S2). This mechanism appears to dominate over the CREB-induced transcriptional activation, since the enzymatic methylation eliminated both induced and basal expression. This notion is supported by further observation that selective partial demethylation of the CRE region did not rescue the expression of the M.SssI methylated reporter constructs (data not shown). In summary, the results demonstrate that methylation and even hemi-methylation of single CpG dinucleotides in critical gene regulatory elements can have substantial negative impacts on the gene expression by directly impeding the transcription factor binding. It is necessary to stress, however, that this effect can only be functionally significant when the surrounding DNA sequence is unmethylated.
The outcome of CRE hydroxymethylation on the gene expression was essentially the same as of methylation (Figures 1–3), thus suggesting that the functional modes of inhibition of the activator-induced gene transcription by these two modifications are similar or common. As for 5-mC, the magnitude of negative effect of single 5-hmC on the gene expression is consistent with the impediment of CREB binding to CRE sequence by the modification. This result suggests that the effects of both 5-mC and 5-hmC are likely mediated by prevention of binding of activatory proteins specific for unmodified CpG rather than by recruitment of co-repressors via a specific interaction with 5-mC or 5-hmC. Such a mechanism is in agreement with proteomics data which identified little overlap between the spectra of specific interactors of 5-mC and 5-hmC, whereas binding of a much larger set of proteins, including several families of transcriptional activators, was disrupted by either modification (22).
The degrees of inhibition of gene expression by single 5-mC and 5-hmC stayed steady over the whole time-course of the gene expression analyses (Figures 2 and 3), suggesting that epigenetic state of the promoter region remained stable over at least 48 hours. In a marked contrast, the expression of constructs containing 5-fC or 5-caC showed strong negative dynamics, indicating a progressive silencing of the affected promoter (Figures 4 and 5). Intriguingly, the spectrum of putative 5-fC and 5-caC readers previously identified by the proteomics approaches includes candidates with potential gene silencing functions such as DNMT1 and the components of the Mi-2/NuRD nucleosome remodeling deacetylase complex (22,39). On the other hand, the enrichment of DNA repair proteins including TDG and MPG in the sets of putative 5-fC and 5-caC readers raises the possibility that proteins bound to products or the DNA repair reactions have been also classified as interactors in these experimental setups.
We have previously reported that excision of various base modifications by the specific DNA glycosylases in concert with the apurinic/apyrimidinic site endonuclease commonly initiates transcriptional silencing (27,29,33). Now we show that TDG clearly contributes to progressive transcriptional silencing induced by 5-caC and 5-fC (Figure 6A), which strongly suggests the key role of BER in this process. Incomplete recovery of the gene expression levels under the conditions of TDG knockdown should likely be attributed to low residual TDG activity in cells, since complete abolition of excision of 5-fC/5-caC by the deoxyribose 2′-fluorination fully restored the expression (Figure 6B and C). Nevertheless, based on these results it still cannot be ruled out that some other DNA glycosylase (or endonuclease) could also contribute to processing of 5-fC and/or 5-caC. Thus, NEIL1, NEIL2 and NEIL3 interact with TET1 and have been implicated in transcriptional activation of TET-oxidised M.SssI-methylated vector DNA in the absence of TDG (40). Even though the evidence is missing that NEIL proteins autonomously excise 5-fC, another study showed that NEIL1 and NEIL2 accelerate the TDG turnover and thus may stimulate excision when TDG is limited (15). Since our findings implicate a BER intermediate in transcriptional silencing induced by 5-fC and 5-caC, it is intriguing to suggest that balance between the activity of individual enzymes within BER or coordination between the BER steps in a given cell type or during a particular differentiation phase may have profound consequences for the regulation of gene expression by these DNA modifications. In our system, both generation of AP site and subsequent strand cleavage by APE1 are required for the gene silencing (Figure 6, Supplementary Figure S6). Taking into account intrinsically low processivity of TDG, it would be important to investigate, whether the switches exist which would halt one of these steps thereby possibly preventing the gene silencing. Such mechanisms could be critical to safeguard the proper gene function following the restoration of unmethylated CpG in the genome.
In summary, our findings have implications for understanding of transcription regulatory functions of hemi-methylated (or hemi-modified by 5-hmC, 5-fC or 5-caC) CpG dinucleotides. Such CpG modifications can arise in cells during DNA demethylation or de novo DNA methylation reactions. The results indicate that 5-mC and 5-hmC act as stable regulatory marks predominantly by decreasing binding of transcriptional activator CREB to the modified target motif. The results further suggest that 5-fC and 5-caC trigger dynamic changes in the promoter activity. Strikingly, in the minimal CRE-regulated promoter employed here, both 5-fC and 5-caC functioned primarily as repressory marks. The impacts of 5-fC and 5-caC on the promoter activity are only partly attributable to the direct impairment of CREB binding and the negative effects on the gene expression strongly advance with time in a TDG-dependent fashion. Extrapolating present results to the situation during active DNA demethylation in cells, we suggest that generation of 5-fC and 5-caC in the regulatory promoter elements and further processing of these modifications by BER by default leads to perpetuation and enhancement of the repressed transcriptional state and that transcriptional re-activation would require additional signals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Bork Lühnsdorf, Bernd Epe, Alexander Ishchenko, Lars Schomacher and Michael Musheev for discussions of the experimental data. The authors are grateful to Bernd Epe and his group members for free access to the FACS instrument and other laboratory facilities.
FUNDING
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) [KH 263/1, KH263/2, Heisenberg Fellowship KH 263/3 to A.K; GRK 2062 Fellowship to M.R.]. Funding for open access charge: DFG, German Research Foundation.
Conflict of interest statement. None declared.
REFERENCES




Comments