Probing the competition between duplex and G-quadruplex/i-motif structures using a conformation-sensitive fluorescent nucleoside probe†
Abstract
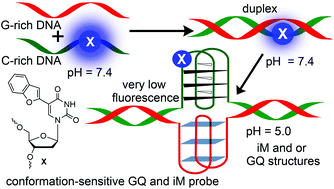
Double-stranded segments of a genome that can potentially form G-quadruplex (GQ) and/or i-motif (iM) structures are considered to be important regulatory elements. Hence, the development of a common probe that can detect GQ and iM structures and also distinguish them from a duplex structure will be highly useful in understanding the propensity of such segments to adopt duplex or non-canonical four-stranded structures. Here, we describe the utility of a conformation-sensitive fluorescent nucleoside analog, which was originally developed as a GQ sensor, in detecting the iM structures of C-rich DNA oligonucleotides (ONs). The analog is based on a 5-(benzofuran-2-yl)uracil scaffold, which when incorporated into C-rich ONs (e.g., telomeric repeats) fluorescently distinguishes an iM from random coil and duplex structures. Steady-state and time-resolved fluorescence techniques enabled the determination of transition pH for the transformation of a random coil to an iM structure. Furthermore, a qualitative understanding on the relative population of duplex and GQ/iM forms under physiological conditions could be gained by correlating the fluorescence, CD and thermal melting data. Taken together, this sensor could provide a general platform to profile double-stranded promoter regions in terms of their ability to adopt four-stranded structures, and also could support approaches to discover functional GQ and iM binders.
- This article is part of the themed collection: Chemical Biology in OBC


 Please wait while we load your content...
Please wait while we load your content...