Potentiometric sensing of nucleic acids using chemically modified nanopores†
Abstract
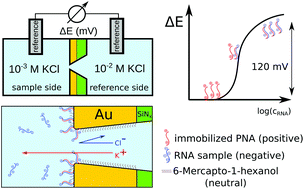
Unlike the overwhelming majority of nanopore sensors that are based on the measurement of a transpore ionic current, here we introduce a potentiometric sensing scheme and demonstrate its application for the selective detection of nucleic acids. The sensing concept uses the charge inversion that occurs in the sensing zone of a nanopore upon binding of negatively charged microRNA strands to positively charged peptide-nucleic acid (PNA) modified nanopores. The initial anionic permselectivity of PNA-modified nanopores is thus gradually changed to cationic permselectivity, which can be detected simply by measuring the nanoporous membrane potential. A quantitative theoretical treatment of the potentiometric microRNA response is provided based on the Nernst–Planck/Poisson model for the nanopore system assuming first order kinetics for the nucleic acid hybridization. An excellent correlation between the theoretical and experimental results was observed, which revealed that the binding process is focused at the nanopore entrance with contributions from both in pore and out of pore sections of the nanoporous membrane. The theoretical treatment is able to give clear guidelines for further optimization of potentiometric nanopore-based nucleic acid sensors by predicting the effect of the most important experimental parameters on the potential response.

 Please wait while we load your content...
Please wait while we load your content...