Functional roles of eriocalyxin B in zebrafish revealed by transcriptome analysis†
Abstract
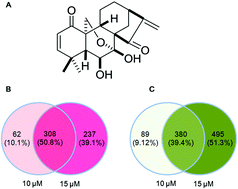
Background. Eriocalyxin B (EriB) is a natural ent-kaurane diterpenoid obtained from Isodon eriocalyx var. laxiflora (family Lamiaceae), which has multiple biological activities (e.g. anti-tumor and anti-inflammatory) via the alteration of gene expression and signaling transduction. Recently, RNA sequencing (RNA-seq) has been developed as a dynamic transcriptome approach to analyze the transcriptional profile and in addition use such gene expression profiles to identify novel candidate genes in a zebrafish model. In the present study, a transcriptome analysis was performed to identify differentially expressed genes (DEGs) in an EriB-exposed zebrafish model. Results. RNA sequencing was conducted on zebrafish embryos after EriB (10 μM and 15 μM) treatment for 72 h. A total of 1570 (405 up-regulated and 1165 down-regulated) and 2511 genes (543 up-regulated and 1968 down-regulated) were identified in the 10 μM and 15 μM groups, respectively. Gene ontology analysis was then performed to elucidate the mechanism of action and effects of EriB. We found that 4 pathways were significantly enriched, which include glutathione metabolism, the metabolism of xenobiotics by cytochrome P450, tight junctions, and phototransduction. The critical transcriptional regulators for the DEGs were also identified by Ingenuity Pathway Analysis after the construction of a protein–protein network, which involves p53, c-myc, binding transcription factor 2, sterol regulatory element binding transcription factor 2, nuclear factor erythroid 2 like 2, and interferon regulatory factor 3. Conclusion. In summary, this is the first study to comprehensively explore the effects of EriB in a zebrafish model using a transcriptome analysis approach. Several important genes with substantial changes in expression levels were discovered. The results of this study will provide insights for the future investigation on the biological activities or toxic effects of EriB.


 Please wait while we load your content...
Please wait while we load your content...