QM/MM simulations indicate that Asp185 is the likely catalytic base in the enzymatic reaction of HIV-1 reverse transcriptase
Abstract
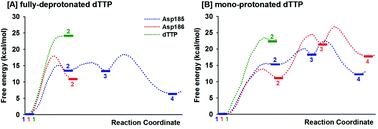
Alternative possible mechanisms of reaction in HIV-1 reverse transcriptase are studied here by QM/MM molecular dynamics umbrella sampling simulations. Two protonation states of the dTTP substrate are tested. Among three different pathways, Asp185 is the probable base for deprotonation of the 3′-primer terminus prior to nucleotide addition on fully-deprotonated dTTP with formation of the stable final product complex of DNAn+1 and PPi. In contrast, the reactions via dTTP or Asp186 as the base show higher energy barriers for either deprotonation or nucleotide addition.

 Please wait while we load your content...
Please wait while we load your content...