Integrative analysis of human omics data using biomolecular networks
Abstract
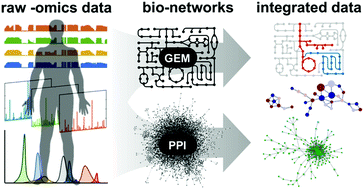
High-throughput ‘-omics’ technologies have given rise to an increasing abundance of genome-scale data detailing human biology at the molecular level. Although these datasets have already made substantial contributions to a more comprehensive understanding of human physiology and diseases, their interpretation becomes increasingly cryptic and nontrivial as they continue to expand in size and complexity. Systems biology networks offer a scaffold upon which omics data can be integrated, facilitating the extraction of new and physiologically relevant information from the data. Two of the most prevalent networks that have been used for such integrative analyses of omics data are genome-scale metabolic models (GEMs) and protein–protein interaction (PPI) networks, both of which have demonstrated success among many different omics and sample types. This integrative approach seeks to unite ‘top-down’ omics data with ‘bottom-up’ biological networks in a synergistic fashion that draws on the strengths of both strategies. As the volume and resolution of high-throughput omics data continue to grow, integrative network-based analyses are expected to play an increasingly important role in their interpretation.

 Please wait while we load your content...
Please wait while we load your content...