KinView: a visual comparative sequence analysis tool for integrated kinome research
Abstract
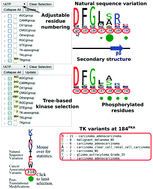
Multiple sequence alignments (MSAs) are a fundamental analysis tool used throughout biology to investigate relationships between protein sequence, structure, function, evolutionary history, and patterns of disease-associated variants. However, their widespread application in systems biology research is currently hindered by the lack of user-friendly tools to simultaneously visualize, manipulate and query the information conceptualized in large sequence alignments, and the challenges in integrating MSAs with multiple orthogonal data such as cancer variants and post-translational modifications, which are often stored in heterogeneous data sources and formats. Here, we present the Multiple Sequence Alignment Ontology (MSAOnt), which represents a profile or consensus alignment in an ontological format. Subsets of the alignment are easily selected through the SPARQL Protocol and RDF Query Language for downstream statistical analysis or visualization. We have also created the Kinome Viewer (KinView), an interactive integrative visualization that places eukaryotic protein kinase cancer variants in the context of natural sequence variation and experimentally determined post-translational modifications, which play central roles in the regulation of cellular signaling pathways. Using KinView, we identified differential phosphorylation patterns between tyrosine and serine/threonine kinases in the activation segment, a major kinase regulatory region that is often mutated in proliferative diseases. We discuss cancer variants that disrupt phosphorylation sites in the activation segment, and show how KinView can be used as a comparative tool to identify differences and similarities in natural variation, cancer variants and post-translational modifications between kinase groups, families and subfamilies. Based on KinView comparisons, we identify and experimentally characterize a regulatory tyrosine (Y177PLK4) in the PLK4 C-terminal activation segment region termed the P+1 loop. To further demonstrate the application of KinView in hypothesis generation and testing, we formulate and validate a hypothesis explaining a novel predicted loss-of-function variant (D523NPKCβ) in the regulatory spine of PKCβ, a recently identified tumor suppressor kinase. KinView provides a novel, extensible interface for performing comparative analyses between subsets of kinases and for integrating multiple types of residue specific annotations in user friendly formats.


 Please wait while we load your content...
Please wait while we load your content...