Structural complexes of the agonist, inverse agonist and antagonist bound C5a receptor: insights into pharmacology and signaling†
Abstract
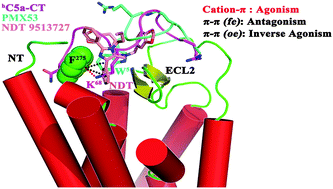
The C5a receptor (C5aR) is a pharmacologically important G-protein coupled receptor (GPCR) that interacts with hC5a, by recruiting both the “orthosteric” sites (site1 at the N-terminus and site2 at the ECS, extra cellular surface) on C5aR in a two site-binding model. However, the complex pharmacological landscape and the distinguishing chemistry operating either at the “orthosteric” site1 or at the functionally important “orthosteric” site2 of C5aR are still not clear, which greatly limits the understanding of C5aR pharmacology. One of the major bottlenecks is the lack of an experimental structure or a refined model structure of C5aR with appropriately defined active sites. The study attempts to understand the pharmacology at the “orthosteric” site2 of C5aR rationally by generating a highly refined full-blown model structure of C5aR through advanced molecular modeling techniques, and further subjecting it to automated docking and molecular dynamics (MD) studies in the POPC bilayer. The first series of structural complexes of C5aR respectively bound to a linear native peptide agonist (hC5a-CT), a small molecule inverse agonist (NDT) and a cyclic peptide antagonist (PMX53) are reported, apparently establishing the unique pharmacological landscape of the “orthosteric” site2, which also illustrates an energetically distinct but coherent competitive chemistry (“cation–π” vs. “π–π” interactions) involved in distinguishing the established ligands known for targeting the “orthosteric” site2 of C5aR. Over a total of 1 μs molecular dynamics (MD) simulation in the POPC bilayer, it is evidenced that while the agonist prefers a “cation–π” interaction, the inverse agonist prefers a “cogwheel/L-shaped” interaction in contrast to the “edge-to-face/T-shaped” type π–π interactions demonstrated by the antagonist by engaging the F2757.28 of the C5aR. In the absence of a NMR or crystallographically guided model structure of C5aR, the computational model complexes not only provide valuable insights for understanding the C5aR pharmacology, but also emerge as a promising platform for the design and discovery of future potential drug candidates targeting the hC5a–C5aR signaling axes.

 Please wait while we load your content...
Please wait while we load your content...