Parameter identification using stochastic simulations reveals a robustness in CD95 apoptotic response†
Abstract
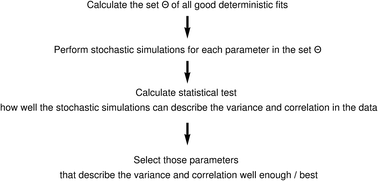
A number of mathematical models of apoptosis generated recently allowed us to understand intrinsic mechanisms of life/death decisions in a cell. Nevertheless, the parameters for the mathematical models are often experimentally difficult to obtain and there is an emerging need for the development of efficient approaches for parameter estimation. In this study we suggest a new method for parameter estimation, which is based on stochastic simulations and can be used when the number of molecules in the system is small. Our approach comprised the following steps: we start from the selection of parameters that lead to a good ordinary differential equation (ODE) fit. We continued by carrying out stochastic simulations for each of these parameters. Comparing the correlation structure of these simulations with the data, we finally could identify the best parameter set. The method was applied for a model of CD95-induced apoptosis, the new best identified parameters fit well to the experimental data. The best parameter set allowed us to get new insights into CD95 apoptosis regulation and can be applied for the comprehensive analysis of other signaling networks. The modeling approach allowed us to get new insights into network regulation, in particular, to identify robustness in CD95 apoptotic response. Taken together, this new method provides valuable predictions and can be applied for the analysis of other signaling networks.

 Please wait while we load your content...
Please wait while we load your content...