Understanding the combinatorial action of transcription factors and microRNA regulation from regions of open chromatin†
Abstract
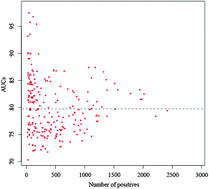
Transcriptional regulatory cascades are always triggered through the combinatorial interplay between transcription factors (TFs) and microRNAs (miRNAs) in eukaryotes. However, it is still a very substantial undertaking to dynamically profile their coordinated actions. In this work, we compared the differences in TFBS numbers between miRNA targets and non-targets, and found that miRNA targets tend to have more TFBS numbers. With the attempt to comprehensively understand the combinatorial action of TF and miRNA regulation from regions of open chromatin, we retrieved recently published DNase I hypersensitive sites (DHSs) across different human cell lines. The result showed that the differences are more statistically significant in DHS regions than non-DHS regions. Next, we trained classifiers for miRNA targets and non-targets. The result showed that TFBSs located in DHS regions achieved a competitive performance when discriminating miRNA targets and non-targets, whereas the performance of classifiers using TFBSs located in non-DHS regions is close to that of a random classifier. After the DHSs were divided into intergenic, transcription start sites (TSSs) and gene body DHS regions based on their genomic locations, only TFBSs located in TSS DHS regions provided a competitive performance. Our results provide us a clue that the coordinated activity of miRNAs and TFs describing the mechanism of gene expression control should be examined in a dynamic perspective.

 Please wait while we load your content...
Please wait while we load your content...