Conformational thermodynamics guided structural reconstruction of biomolecular fragments†
Abstract
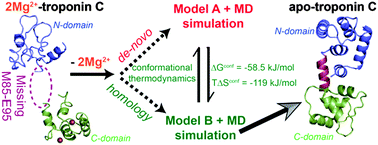
Computational prediction of structure for macromolecular fragments is a formidable challenge. Here we show that the differences in conformational thermodynamics, computed using the equilibrium distribution of dihedral angles from molecular dynamics simulation, can identify the better model for the missing residues in the metal ion free (apo) skeletal muscle Troponin C (TnC). We use the model to understand Troponin I interaction with calcium (Ca2+) ion bound TnC. Our method to compare conformational thermodynamics between different models can be easily generalized to any macromolecule to understand the structure and function even if experimental structures are not resolved.
- This article is part of the themed collection: Chemical Biology in Molecular BioSystems

 Please wait while we load your content...
Please wait while we load your content...