The effect of sequence context on the activity of cytosine DNA glycosylases†
Abstract
We have prepared single (N204D) and double (N204D:L272A) mutants of human uracil DNA glycosylase (hUDG), generating two cytosine DNA glycosylases (hCDG and hCYDG). Both these enzymes are able to excise cytosine (but not 5-methylcytosine), when this base is part of a mismatched base pair. hCDG is more active than the equivalent E. coli enzyme (eCYDG) and also has some activity when the cytosine is paired with guanine, unlike eCYDG. hCDG also has some activity against single stranded DNA, while having poor activity towards an unnatural base pair that forces the cytosine into an extrahelical conformation (in contrast to eCYDG for which a bulky base enhances the enzyme's activity). We also examined how sequence context affects the activity of these enzymes, determining the effect of flanking base pairs on cleavage efficiency. An abasic site or a hexaethylene glycol linker placed opposite the target cytosine, also causes an increase in activity compared with an AC mismatch. Flanking an AC mismatch with GC base pairs resulted in a 100-fold decrease in excision activity relative to flanking AT base pairs and the 5′-flanking base pair had a greater effect on the rate of cleavage. However, this effect is not simply due to the stability of the flanking base pairs as adjacent GT mismatches also produce low cleavage efficiency.
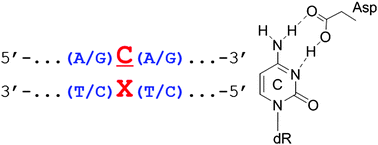
- This article is part of the themed collection: Chemical Biology in Molecular BioSystems

 Please wait while we load your content...
Please wait while we load your content...