Improving gene regulatory network inference using network topology information†
Abstract
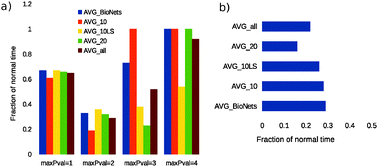
Inferring the gene regulatory network (GRN) structure from data is an important problem in computational biology. However, it is a computationally complex problem and approximate methods such as heuristic search techniques, restriction of the maximum-number-of-parents (maxP) for a gene, or an optimal search under special conditions are required. The limitations of a heuristic search are well known but literature on the detailed analysis of the widely used maxP technique is lacking. The optimal search methods require large computational time. We report the theoretical analysis and experimental results of the strengths and limitations of the maxP technique. Further, using an optimal search method, we combine the strengths of the maxP technique and the known GRN topology to propose two novel algorithms. These algorithms are implemented in a Bayesian network framework and tested on biological, realistic, and in silico networks of different sizes and topologies. They overcome the limitations of the maxP technique and show superior computational speed when compared to the current optimal search algorithms.


 Please wait while we load your content...
Please wait while we load your content...