In silico identification of novel kinase inhibitors targeting wild-type and T315I mutant ABL1 from FDA-approved drugs†
Abstract
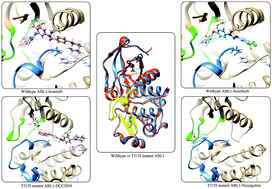
The constitutively active fusion protein BCR-ABL1 is the major cause of chronic myeloid leukemia (CML), and selective inhibition of ABL1 is a promising approach for the treatment of CML. Reported drugs worked well in clinical practice, such as imatinib, dasatinib, nilotinib and bosutinib. However, resistance arises due to ABL1 mutation in patients, especially the T315I gate-keeper mutation. Thus, wide spectrum drugs targeting ABL1 are urgently needed. In order to screen potential drugs targeting wild-type ABL1 and T315I mutant ABL1, 1408 FDA approved small molecule drugs were subjected to molecular docking. With subsequent molecular dynamic (MD) simulation and MM/GBSA binding free energy calculation and energy decomposition, we identified chlorhexidine and sorafenib as potential “new use” drugs targeting wild-type ABL1, while nicergoline and plerixafor targeted T315I ABL1. Meanwhile, we also found that residues located in the ATP-binding site and A-loop motif played key roles in drug discovery towards ABL1. These findings may not only serve as a paradigm for the repositioning of existing approved drugs, but also instill new vitality to ABL1-targeted anti-CML therapeutics.

 Please wait while we load your content...
Please wait while we load your content...