Role of cytoskeletal proteins in cerebral cavernous malformation signaling pathways: a proteomic analysis†
Abstract
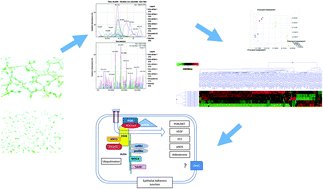
Three genetic mutations were found to cause cerebral cavernous malformation (CCM), a vascular anomaly predisposing affected individuals to hemorrhagic stroke. These CCM proteins function together as a protein complex in the cell. Loss of expression of each CCM gene results in loss of in vitro endothelial tube formation. Label-free differential protein expression analysis using multidimensional liquid chromatography/tandem mass spectrometry (2D-LC-MS/MS) was applied to explore the proteomic profile for loss of each CCM gene expression in mouse endothelial stem cells (MEES) compared to mock shRNA and no shRNA control cell-lines. Differentially expressed proteins were identified (p < 0.05). 120 proteins were differentially expressed among the cell-lines. Principal component analysis and cluster analysis show the effects of individual knockdown. In all knockdown cell-lines, altered expression of cytoskeletal proteins is the most common. While all CCM mutations result in similar pathology, different CCM mutations have their own distinct pathogenesis in cell signaling.

 Please wait while we load your content...
Please wait while we load your content...