Power graph compression reveals dominant relationships in genetic transcription networks†
Abstract
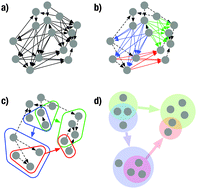
We introduce a framework for the discovery of dominant relationship patterns in transcription networks, by compressing the network into a power graph with overlapping power nodes. Our application of this approach to the transcription networks of S. cerevisiae and E. coli, paired with GO term enrichment analysis, provides a highly informative overview of the most prominent relationships in the gene regulatory networks of these two organisms.

 Please wait while we load your content...
Please wait while we load your content...