Proteomic analysis of formalin-fixed paraffin-embedded colorectal cancer tissue using tandem mass tag protein labeling†
Abstract
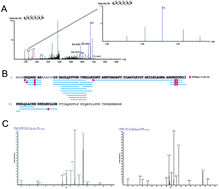
In clinical research, repositories of biological samples form a rich source of clinical material for biomarker studies. Banked material, however, is often not stored in optimal conditions regarding the technology used for biomarker research. A case in point is formalin-fixed paraffin-embedded (FFPE) tissue that could be used to obtain large cohorts of samples over a short period of time, as these tissues are routinely prepared for pathological analysis. However, in the context of mass spectrometry based peptide-centric proteomics, protein extraction and identification can be hampered by formalin-induced crosslinking. Furthermore, the molecular formalin crosslinks might be entangled differently across various samples, making it more difficult to reproducibly extract the same proteins from different samples. In this study, we establish the crosslink variability using Tandem Mass Tag (TMT) protein labeling followed by digestion, separation, identification and quantification of proteins extracted from FFPE colorectal cancer and paired healthy tissues. Moreover, by applying de novo interpretation of tandem mass spectra and subsequent analysis by Peaks PTM, unspecified modifications could be elucidated, leading to increased protein and proteome coverage. This approach might be useful for future FFPE proteomics studies.

 Please wait while we load your content...
Please wait while we load your content...