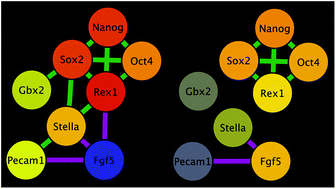
Analysis of transcription at the level of single cells in prokaryotes and eukaryotes has revealed the existence of heterogeneities in the expression of individual genes within genetically homogeneous populations. This variation is an emerging hallmark of populations of Embryonic Stem (ES) cells and has been ascribed to the stochasticity associated with the biochemical events that mediate gene expression. It has been suggested that these heterogeneities play a role in the maintenance of pluripotency. However, for the most part, studies have focused on individual genes in large cell populations. Here we use an existing dataset on the expression of eight genes involved in pluripotency in eighty-three ES cells to create Gene Regulatory Networks (GRNs) at the single cell level. We observe widespread heterogeneities in the expression of the eight genes, but analysis of correlations within individual cells reveals three distinct classes centered on the expression of Nanog, a marker of pluripotency, and Fgf5, a gene associated with differentiation: high levels of Nanog and low levels of Fgf5, low levels of Nanog and high levels of Fgf5, and low levels of both. Each of these classes is associated with a collection of active sub-networks, with differing degrees of connectivity between their elements, which define a cellular state: self-renewal, primed for differentiation or transition between the two. Though every cell should be governed by the same network, the active sub-networks may emerge due to considerations such as variation in (i) the expression level of active transcription factors (e.g. through post-translational modification or ligand/co-factor availability) or (ii) access to the target gene locus (e.g. via changes in chromatin status or epigenetic modifications). We conclude that heterogeneities in gene expression should not be interpreted as representing different states of a single unique network, but as a reflection of the activity of different sub-networks in sub-populations of cells.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...