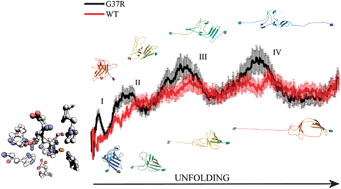
Although the molecular determinants of Familial Amyotrophic Lateral Sclerosis (FALS) are still largely unknown, previous studies have demonstrated that aggregation of Cu, Zn superoxide dismutase (SOD1) mutants may play a causative role in FALS. It has been proposed that this pathogenic process occurs via a multi-step pathway involving metal loss, dimer dissociation and assembly of misfolded apo-monomers. The G37R, one of the many SOD1 mutations known to be associated to FALS, is difficult to be reconciled with this model because it is located far from the metal sites and the monomer–monomer interface. Consequently, an inspection of all the steps involved in G37R SOD1 misfolding is expected to provide hints in the understanding of the molecular basis of the disease. To this aim, an array of different computational strategies—i.e. Thermodynamic Integration (TI), implicit solvent Constant Temperature Molecular Dynamics (CTMD) and Steered Molecular Dynamics (SMD)—have been applied on the G37R SOD1 mutant. A comparison with parallel studies carried out for the Wild Type (WT) SOD1 pointed out that the mutation decreases the affinity of the protein for the Cu(II) ion. Implicit solvents MD simulations performed on the two apo proteins revealed that in the mutant SOD1 a novel, stable H-bond network involving Arg37, Lys91, Lys36 and Leu38 is created thus confirming a pivotal role of this region in driving the biophysical properties of the entire protein. Finally, the presence of energetic “traps” in the force vs. elongation curves of G37R SOD1 is an indicator of the existence of intermediate states along the unfolding pathway which may lead to abnormal conformers. Our results support a general theory suggesting that the two major hypotheses regarding mutant SOD1 toxicity, i.e. aberrant copper redox chemistry and SOD1 misfolding are causally linked. In fact it is shown that the G37R mutation, although located far away the active site, may induce subtle modification in SOD1 leading to the loosening of metal binding and to the formation of metastable intermediate states along the unfolding pathway.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...