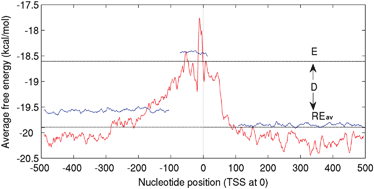
The rapid increase in genome sequence information has necessitated the annotation of their functional elements, particularly those occurring in the non-coding regions, in the genomic context. Promoter region is the key regulatory region, which enables the gene to be transcribed or repressed, but it is difficult to determine experimentally. Hence an in silico identification of promoters is crucial in order to guide experimental work and to pin point the key region that controls the transcription initiation of a gene. In this analysis, we demonstrate that while the promoter regions are in general less stable than the flanking regions, their average free energy varies depending on the GC composition of the flanking genomic sequence. We have therefore obtained a set of free energy threshold values, for genomic DNA with varying GC content and used them as generic criteria for predicting promoter regions in several microbial genomes, using an in-house developed tool ‘PromPredict’. On applying it to predict promoter regions corresponding to the 1144 and 612 experimentally validated TSSs in E. coli (50.8% GC) and B. subtilis (43.5% GC) sensitivity of 99% and 95% and precision values of 58% and 60%, respectively, were achieved. For the limited data set of 81 TSSs available for M. tuberculosis (65.6% GC) a sensitivity of 100% and precision of 49% was obtained.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...