Kinetic analysis methods applied to single motor protein trajectories†
Abstract
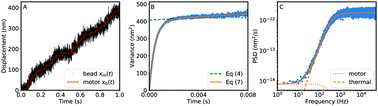
Molecular motors convert chemical or electrical energy into mechanical displacement, either linear or rotary. Under ideal circumstances, single-molecule measurements can spatially and temporally resolve individual steps of the motor, revealing important properties of the underlying mechanochemical process. Unfortunately, steps are often hard to resolve, as they are masked by thermal noise. In such cases, details of the mechanochemistry can nonetheless be recovered by analyzing the fluctuations in the recorded traces. Here, we expand upon existing statistical analysis methods, providing two new avenues to extract the motor step size, the effective number of rate-limiting chemical states per translocation step, and the compliance of the link between the motor and the probe particle. We first demonstrate the power and limitations of these methods using simulated molecular motor trajectories, and we then apply these methods to experimental data of kinesin, the bacterial flagellar motor, and F1-ATPase.


 Please wait while we load your content...
Please wait while we load your content...