Characterization of the structural ensembles of p53 TAD2 by molecular dynamics simulations with different force fields†
Abstract
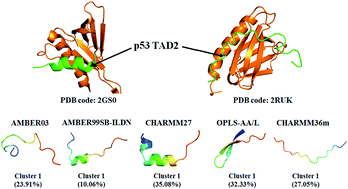
Intrinsically disordered regions (IDRs) or proteins (IDPs), which play crucial biological functions in essential biological processes of life, do not have well-defined secondary or tertiary structures when isolated in solution. The highly dynamic properties and conformational heterogeneity of IDPs make them challenging to study with traditional experimental techniques. As a powerful complementary tool for experiments, all-atom molecular dynamics simulation can obtain detailed conformational information on IDPs, but the limitation of force field accuracy is a challenge for reproducing IDP conformers. Here, we compared five empirical all-atom force fields AMBER03, AMBER99SB-ILDN, CHARMM27, OPLS-AA/L and CHARMM36m in modeling the conformational ensembles of wild-type peptide TAD2(41–62) from the human p53 tumor suppressor. Our results show that for the model peptide, the newest force field CHARMM36m produces more expanded coil ensemble followed by AMBER99SB-ILDN; CHARMM27 displays a predominant propensity for a helical structure; whereas OPLS-AA/L exhibits a apparent preference for a β-sheet structure and yields the most compact conformation. In the comparison of the simulated dimensions with theoretical prediction and the back-calculated chemical shifts with experimental measurements, AMBER99SB-ILDN gives a more consistent agreement than the other force fields. In addition, the region from residues 47 to 55, which commonly forms an amphipathic α-helix upon binding target proteins according to experimental observation, could form a helical structure with a different probability population in our simulations with different force fields. This implies that the binding process might be conducted by, or partly by “conformation selection” for this peptide. This work indicates that force field development for modeling general IDPs accurately has a long way to go, and more detailed experimental data of IDPs are also in demand.


 Please wait while we load your content...
Please wait while we load your content...