The folding mechanism and key metastable state identification of the PrP127–147 monomer studied by molecular dynamics simulations and Markov state model analysis†
Abstract
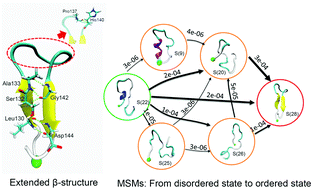
The structural transition of prion proteins from a native α-helix (PrPC) to a misfolded β-sheet-rich conformation (PrPSc) is believed to be the main cause of a number of prion diseases in humans and animals. Understanding the molecular basis of misfolding and aggregation of prion proteins will be valuable for unveiling the etiology of prion diseases. However, due to the limitation of conventional experimental techniques and the heterogeneous property of oligomers, little is known about the molecular architecture of misfolded PrPSc and the mechanism of structural transition from PrPC to PrPSc. The prion fragment 127–147 (PrP127–147) has been reported to be a critical region for PrPSc formation in Gerstmann–Straussler–Scheinker (GSS) syndrome and thus has been used as a model for the study of prion aggregation. In the present study, we employ molecular dynamics (MD) simulation techniques to study the conformational change of this fragment that could be relevant to the PrPC–PrPSc transition. Employing extensive replica exchange molecular dynamics (REMD) and conventional MD simulations, we sample a huge number of conformations of PrP127–147. Using the Markov state model (MSM), we identify the metastable conformational states of this fragment and the kinetic network of transitions between the states. The resulting MSM reveals that disordered random-coiled conformations are the dominant structures. A key metastable folded state with typical extended β-sheet structures is identified with Pro137 being located in a turn region, consistent with a previous experimental report. Conformational analysis reveals that intrapeptide hydrophobic interaction and two key residue interactions, including Arg136–His140 and Pro137–His140, contribute a lot to the formation of ordered extended β-sheet states. However, network pathway analysis from the most populated disordered state indicates that the formation of extended β-sheet states is quite slow (at the millisecond level), as large structural rearrangement is needed from disordered states. We speculate that the formation process of the extended β-sheet folded states may represent an important event during the early formation of prion oligomers and the results of our study provide insights into the molecular details of the early stage of prion aggregation.


 Please wait while we load your content...
Please wait while we load your content...