In silico analysis of interaction pattern switching in ligand⋯receptor binding in Golgi α-mannosidase II induced by the protonated states of inhibitors†
Abstract
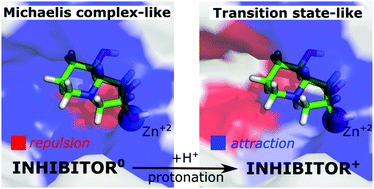
Golgi α-mannosidase II (GM) is a pharmaceutical target for the design of inhibitors with anticancer activity. The known potent GM inhibitors undergo complex interactions with Zn2+ ions and the active-site amino acids, many of which contain ionisable functional groups. Herein, the physical insight into the ligand⋯receptor interactions has been provided based on energy decomposition techniques: SAPT (symmetry adapted perturbation theory) and FMO-PIEDA (fragment molecular orbital-pair interaction energy decomposition analysis) for a large GM active-site cluster. Protonation-dependent molecular recognition in Golgi α-mannosidase was demonstrated for five inhibitors, mannose, and its transition state. Zn2+ ion and Asp472 induce the key interactions with the deprotonated inhibitors (bearing an amino group in the neutral state), followed by Asp92 and Asp341. This interaction pattern is consistent for all the studied inhibitors and is similar to the interaction pattern of the enzyme native substrate – mannose. The interactions with Zn2+ ion become repulsive for the protonated states of the inhibitors (bearing an amino group with +1 charge) and the mannosyl transition state. The importance of Asp92 and Asp204 considerably increases, while the interactions with Asp472 and Asp341 are slightly modified. The interaction pattern for the protonated ligands seems to have an oxocarbenium transition state-like character, rather than a Michaelis complex of GM. The electrostatic interactions with amino acids coordinating zinc ion are of key importance for both the neutral and protonated states of the inhibitors. The ligand's diol group has a dual role as an electron donor, coordinating zinc ion, and as an electron acceptor, interacting with Asp92 and Asp472 via strong hydrogen bonds. This interaction pattern is an essential structural feature of the potent GM inhibitors, which is consistent with the experimental findings. Based on the calculations, either the protonated or deprotonated state of the ligand may be the active form of the GM inhibitor, exhibiting different interacting patterns.


 Please wait while we load your content...
Please wait while we load your content...