Biophysical evaluation of protein structural flexibility for ligand biorecognition in solid solution†
Abstract
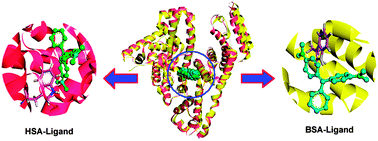
Ligand–protein recognition is increasingly explored through different biophysical approaches, but the effects of protein flexibility are frequently neglected in many applications. The main goal of this contribution is to comparatively study the flexibility and dynamics of homologous model proteins, i.e. human serum albumin/bovine serum albumin (HSA/BSA), in biopolymer–ligand recognition by using computational chemistry based on multispectroscopic data. The results of spectroscopic analyses showed that the recognition of a ligand by a protein formed a noncovalent adduct with a stoichiometry of 1 : 1, and a ligand was situated stably at the subdomain IIA of protein, which corroborates that time-resolved fluorescence displaying the autoregulation/transition of protein conformation occurred and this phenomenon has further been proved by circular dichroism. In light of molecular docking and molecular dynamics simulations, we may confirm that the overall stability of proteins strengthens facilely as the recognition ability of homologous proteins for ligands grows larger. Conversely, if the recognition capability is relatively small, the stability of some amino acid residues can also be enhanced, but the stability of integrally homologous proteins (complexed proteins) did not increase significantly. Furthermore, via careful examination of free energy decomposition, we could find that the electrostatic interactions in the HSA–ligand are pronouncedly greater than the BSA–ligand system; however, the van der Waals forces in the BSA–ligand have the most remarkable energy contributions to the protein–ligand reaction. This suggests that the interaction energy of the two biological systems possesses quite notable discrepancies. Probably the results obtained herein would help in revealing the influences of inherent structural features such as flexibility and dynamics of proteins on biomacromolecule–ligand recognition.

 Please wait while we load your content...
Please wait while we load your content...