Protein–ligand docking using fitness learning-based artificial bee colony with proximity stimuli†
Abstract
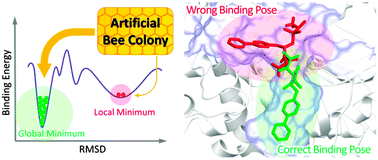
Protein–ligand docking is an optimization problem, which aims to identify the binding pose of a ligand with the lowest energy in the active site of a target protein. In this study, we employed a novel optimization algorithm called fitness learning-based artificial bee colony with proximity stimuli (FlABCps) for docking. Simulation results revealed that FlABCps improved the success rate of docking, compared to four state-of-the-art algorithms. The present results also showed superior docking performance of FlABCps, in particular for dealing with highly flexible ligands and proteins with a wide and shallow binding pocket.

 Please wait while we load your content...
Please wait while we load your content...