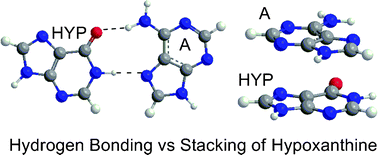
The hydrogen-bonding and stacking interactions of hypoxanthine, a potential universal nucleobase, were calculated using a variety of methodologies (CCSD(T), MP2, B3LYP, PWB6K, AMBER). All methods predict that the hydrogen-bonding interaction in the hypoxanthine–cytosine pair is approximately 25 kJ mol−1 stronger than that in the other dimers. Although the calculations support suggestions from experiments that hypoxanthine preferentially binds with cytosine, the trend in the calculated hydrogen-bond strengths for the remaining natural nucleobases do not show a strong correlation with the experimentally predicted binding preferences. However, our calculations suggest that the stacking interactions of hypoxanthine are similar in magnitude to the hydrogen-bonding interactions at all levels of theory (with the exception of B3LYP, which incorrectly predicts stacked dimers to be unstable). Therefore, stacking interactions should also be considered when analyzing the stability of DNA helices containing hypoxanthine and the use of larger models that account for both hydrogen-bonding and stacking within DNA duplexes will likely result in better agreement with experimental observations. For the majority of the dimers, PWB6K and AMBER provide reasonable binding strengths at reduced computational costs, and therefore will be useful techniques for considering larger models.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...