DNA display of folded RNA libraries enabling RNA-SELEX without reverse transcription†
Abstract
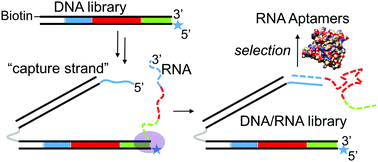
A method for the physical attachment of folded RNA libraries to their encoding DNA is presented as a way to circumvent the reverse transcription step during systematic evolution of RNA ligands by exponential enrichment (RNA-SELEX). A DNA library is modified with one isodC base to stall T7 polymerase and a 5′ “capture strand” which anneals to the nascent RNA transcript. This method is validated in a selection of RNA aptamers against human α-thrombin with dissociation constants in the low nanomolar range. This method will be useful in the discovery of RNA aptamers and ribozymes containing base modifications that make them resistant to accurate reverse transcription.


 Please wait while we load your content...
Please wait while we load your content...