MALDI-ion mobility mass spectrometry of lipids in negative ion mode†
Abstract
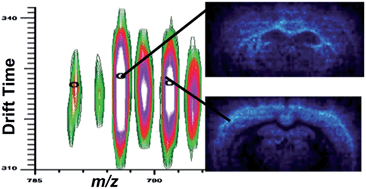
Profiling and imaging using MALDI mass spectrometry (MS) allows detection and localization of biomolecules in tissue, of which lipids are a major component. However, due to the in situ nature of this technique, complexity of tissue and need for a chemical matrix, the recorded signal is complex and can be difficult to assign. Ion mobility adds a dimension that provides coarse shape information, separating isobaric lipids, peptides, and oligonucleotides along distinct familial trend lines before mass analysis. Previous work using MALDI-ion mobility mass spectrometry to analyze and image lipids has been conducted mainly in positive ion mode, although several lipid classes ionize preferentially in negative ion mode. This work highlights recent data acquired in negative ion mode to detect glycerophosphoethanolamines (PEs), glycerophosphoserines (PSs), glycerophosphoglycerols (PGs), glycerolphosphoinositols (PIs), glycerophosphates (PAs), sulfatides (STs), and gangliosides from standard tissue extracts and directly from mouse brain tissue. In particular, this study focused on changes in ion mobility based upon lipid head groups, composition of radyl chain (# of carbons and double bonds), diacyl versus plasmalogen species, and hydroxylation of species. Finally, a MALDI-ion mobility imaging run was conducted in negative ion mode, resulting in the successful ion mapping of several lipid species.

 Please wait while we load your content...
Please wait while we load your content...