Unraveling the inhibitory potential of Rosetta designed de novo cyclic peptides on PARP7 through molecular dynamics simulations
Abstract
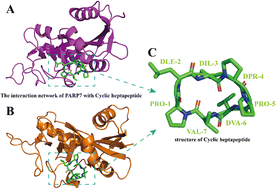
Poly adenosine diphosphate (ADP)-ribose polymers (PARPs) are enzymes that catalyze ADP-ribosylation and regulate fundamental cellular processes in the body. PARP inhibitors are utilized as anticancer agents that target poly(ADP-ribose) polymerase, among which PARP7 and PARP12 are essential members of the PARP family; both are monoPARP and have excellent prospects as antitumor drug targets. The current PARP family inhibitors consist primarily of small molecule drugs, which display high toxicity and low specificity, necessitating cautious usage. Therefore, the current study utilizes computational structural biology approaches to discover inhibitors with enhanced specificity, safety, and affinity toward the target. First, the Rosetta software is used for predicting protein structures and denovo design of cyclic peptides, followed by individually docking numerous cyclic peptides into PARP7 and PARP12 active sites using the ZDOCK server. Finally, molecular dynamics (MD) simulations were conducted on the docked complexes using GROMACS_2020 to evaluate the stability of newly designed cyclic peptides and compute energy scorings. As a result, we identified a cyclic heptapeptide with a significant binding affinity to PARP7, establishing several hydrogen bonds and having high stability with significant binding scores. This analysis highlights the application of MD simulations in the drug design of PARP7 inhibitors, and it also offers a promising starting point for the further development of PARP7 inhibitors.


 Please wait while we load your content...
Please wait while we load your content...