Emulating clinical pressure waveforms in cell culture using an Arduino-controlled millifluidic 3D-printed platform for 96-well plates
Abstract
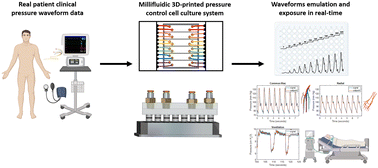
High blood pressure is the primary risk factor for heart disease, the leading cause of death globally. Despite this, current methods to replicate physiological pressures in vitro remain limited in sophistication and throughput. Single-chamber exposure systems allow for only one pressure condition to be studied at a time and the application of dynamic pressure waveforms is currently limited to simple sine, triangular, or square waves. Here, we introduce a high-throughput hydrostatic pressure exposure system for 96-well plates. The platform can deliver a fully-customizable pressure waveform to each column of the plate, for a total of 12 simultaneous conditions. Using clinical waveform data, we are able to replicate real patients' blood pressures as well as other medically-relevant pressures within the body and have assembled a small patient-derived waveform library of some key physiological locations. As a proof of concept, human umbilical vein endothelial cells (HUVECs) survived and proliferated for 3 days under a wide range of static and dynamic physiologic pressures ranging from 10 mm Hg to 400 mm Hg. Interestingly, pathologic and supraphysiologic pressure exposures did not inhibit cell proliferation. By integrating with, rather than replacing, ubiquitous lab cultureware it is our hope that this device will facilitate the incorporation of hydrostatic pressure into standard cell culture practice.


 Please wait while we load your content...
Please wait while we load your content...