Charting and tracking the evolution of the SARS CoV-2 coronavirus variants of concern with protein mass spectrometry†
Abstract
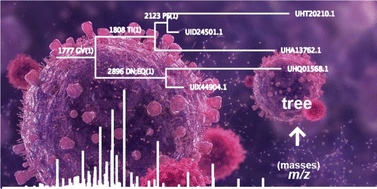
The evolution of the SARS-CoV2 coronavirus spike S-protein is studied using a mass spectrometry based protein phylogenetic approach. A study of a large dataset comprising sets of peptide masses derived from over 3000 proteins of the SARS-CoV2 virus shows that the approach is capable of resolving and correctly displaying the evolution of the major variants of concern. Using these numerical datasets, through a pairwise comparison of sets of proteolytic peptide masses for each protein, the tree is built without the need for the sequence data itself or any sequence alignment. In the same analysis, single point mutations are calculated from peptide mass differences of different protein sets and these are displayed at the branch nodes on the tree. The tree topology is found to be consistent with that generated using conventional sequence-based phylogenetics by a manual visualisation and using a tree comparison algorithm. The mass tree resolves major variants of the virus and displays non-synonymous mutations, calculated based on the mass data alone, on the tree that enable protein evolution to be charted and tracked along interconnected branches. Tracking the evolution of the SARS-CoV2 coronavirus S-protein is of particular importance given its role in the attachment of the virus to host cells ahead of viral replication.


 Please wait while we load your content...
Please wait while we load your content...